BLH3 Regulates the ABA Pathway and Lignin Synthesis Under Salt Stress in Lilium pumilum
- PMID: 40573849
- PMCID: PMC12196856
- DOI: 10.3390/plants14121860
BLH3 Regulates the ABA Pathway and Lignin Synthesis Under Salt Stress in Lilium pumilum
Abstract
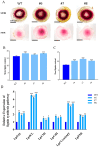
BEL1-like homeodomain protein 3 (BLH3) plays a crucial role in plant development. However, its involvement in the salt stress response has not been studied. In this study, we investigated the molecular mechanism underlying the response of LpBLH3 to salt stress in Lilium pumilum (L. pumilum) using various techniques, including quantitative PCR (RT-qPCR), determination of physiological indices of plant after Saline-Alkali stress, yeast two-hybrid screening, luciferase complementation imaging (LCI), and chromosome walking to obtain the promoter sequence, analyzed by PlantCARE, electrophoretic mobility shift assay (EMSA), and then dual-luciferase reporter assay(LUC). RT-qPCR analysis revealed that LpBLH3 is most highly expressed in the leaves of L. pumilum. The expression of LpBLH3 peaks at 24 or 36 h in the leaves under different saline stress. Under various treatments, compared to the wild type (WT), the LpBLH3 overexpression lines exhibited less chlorosis and leaf curling and stronger photosynthesis. The overexpression of LpBLH3 can enhance lignin accumulation in root and stem by positively modulating the expression of crucial genes within the lignin biosynthesis pathway. Y2H and LCI analyses demonstrated that LpBLH3 interacts with LpKNAT3. Additionally, EMSA and LUC analyses confirmed that LpBLH3 can bind to the promoter of LpABI5 and upregulate the expression of ABI5 downstream genes (LpCAT1/LpATEM/LpRD29B). In summary, LpBLH3 enhances the plant's salt tolerance through the ABA pathway and lignin synthesis. This study can enrich the functional network of the BLH transcription factor family, obtain Lilium pumilum lines with good saline-alkali resistance, expand the planting area of Lilium pumilum, and improve its medicinal and ornamental values. Additionally, the functional analysis of the BLH transcription factor family provides new insights into how crops adapt to the extreme growth environment of saline-alkali soils.
Keywords: ABA pathway; BLH3; Lilium pumilum; lignin content; salt stress.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Wang J. How saline-alkaline land transforms into “black soil”: Placing equal emphasis on improvement and cultivation. The People’s Daily. Nov 1, 2023.
-
- Yao D., Wu J., Hu Z., Bai B., Zhuang W., Li J., Deng Q. Physiological mechanisms and breeding strategies for saline-alkali tolerance in rice. Hybrid Rice. 2019;34:1–7. doi: 10.16267/j.cnki.1005-3956.20180426.130. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

