Mucosal and Serum Neutralization Immune Responses Elicited by COVID-19 mRNA Vaccination in Vaccinated and Breakthrough-Infection Individuals: A Longitudinal Study from Louisville Cohort
- PMID: 40573890
- PMCID: PMC12197714
- DOI: 10.3390/vaccines13060559
Mucosal and Serum Neutralization Immune Responses Elicited by COVID-19 mRNA Vaccination in Vaccinated and Breakthrough-Infection Individuals: A Longitudinal Study from Louisville Cohort
Abstract
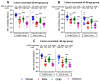
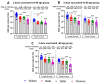
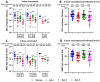
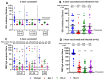
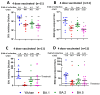
Background/Objectives: The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus type-2 (SARS-CoV-2), has resulted in 777 million cases worldwide. Various vaccines have been approved to control the spread of COVID-19, with mRNA vaccines (Pfizer and Moderna) being widely used in the USA. We conducted a prospective longitudinal study to analyze the immune response elicited by two/three and four doses of monovalent mRNA vaccines in both vaccinated individuals and those who experienced breakthrough infections. Participants were stratified into different age groups: 18-40, 41-60, and over 60 years. Methods: We assessed cross-variant neutralization responses in two cohorts-Cohort I: n = 167 (serum), Cohort II: n = 92 (serum and nasal swab) samples-using infectious virus microneutralization assay (MN) and antibody (IgG or IgA) binding ELISA titers to the spike protein receptor binding domain (RBD). Samples were collected from the Louisville Metro-Jefferson County Co-Immunity Project, a federally funded, population-based study for the surveillance of SARS-CoV-2 in Jefferson County, Kentucky during 2020-2022, involving both health care workers and a local community. Results: Individuals who received two doses of the mRNA vaccine exhibited reduced neutralization against Beta, Delta, and Omicron BA.1 variants compared to wildtype Wuhan, with further decline observed six months post-booster vaccination. However, individuals who experienced natural COVID-19 infection (breakthrough) after receiving two vaccine doses showed enhanced neutralization and antibody responses, particularly against Omicron BA.1. Following the 3rd dose, antibodies and neutralization responses were restored. Among triple-vaccinated individuals, reduced neutralization was observed against Omicron variants BA.1, BA.5, and BA.2 compared to Wuhan. Neutralization responses were better against BA.2 variant compared to BA.1 and BA.5. However, individuals who received three doses of vaccine and experienced a breakthrough infection (n = 45) elicited significantly higher neutralizing antibodies responses against all Omicron subvariants compared to vaccinated individuals. Interestingly, nasal swab samples collected from volunteers with breakthrough infection showed significantly elevated spike-reactive mucosal IgA antibodies and enhanced cross neutralization against BA.1, BA.2, and BA.5 compared to individuals who received only three vaccine doses. Conclusions: mRNA vaccination elicits a strong systemic immune response by boosting serum neutralizing antibodies (NAb), although this protection wanes over time, allowing new variants to escape neutralization. Breakthrough individuals have extra enrichment in nasal NAb offering protection against emerging variants. This longitudinal immune profiling underscores the strengthening of pandemic preparedness and supports the development of durable mucosal vaccines against respiratory infectious disease.
Keywords: COVID-19; IgA and IgG; Omicron; antibodies; breakthrough infections; hybrid immunity; mRNA vaccine; microneutralization (MN); mucosal response; systemic response.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
COVID-19 Vaccines.2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. 2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. PMID: 33355732 Free Books & Documents. Review.
-
Safety and immunogenicity of a modified mRNA-lipid nanoparticle vaccine candidate against COVID-19: Results from a phase 1, dose-escalation study.Hum Vaccin Immunother. 2024 Dec 31;20(1):2408863. doi: 10.1080/21645515.2024.2408863. Epub 2024 Oct 18. Hum Vaccin Immunother. 2024. PMID: 39422261 Free PMC article. Clinical Trial.
-
Determining the Time of Booster Dose Based on the Half-Life and Neutralization Titers against SARS-CoV-2 Variants of Concern in Fully Vaccinated Individuals.Microbiol Spectr. 2023 Aug 17;11(4):e0408122. doi: 10.1128/spectrum.04081-22. Epub 2023 Jul 10. Microbiol Spectr. 2023. PMID: 37428104 Free PMC article.
-
Elicitation of neutralizing antibodies and IgG4 subclass switching following booster vaccination with ancestral COVID-19 mRNA vaccines does not reduce breakthrough infections.Hum Vaccin Immunother. 2025 Dec;21(1):2547517. doi: 10.1080/21645515.2025.2547517. Epub 2025 Aug 14. Hum Vaccin Immunother. 2025. PMID: 40812315 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
References
-
- Alharbi A.M., Rabbani S.I., Halim Mohamed A.A., Almushayti B.K., Aldhwayan N.I., Almohaimeed A.T., Alharbi A.A., Alharbi N.S., Asdaq S.M.B., Alamri A.S., et al. Analysis of potential risk factors associated with COVID-19 and hospitalization. Front. Public Health. 2022;10:921953. doi: 10.3389/fpubh.2022.921953. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

