Evolving SARS-CoV-2 Vaccines: From Current Solutions to Broad-Spectrum Protection
- PMID: 40573967
- PMCID: PMC12197680
- DOI: 10.3390/vaccines13060635
Evolving SARS-CoV-2 Vaccines: From Current Solutions to Broad-Spectrum Protection
Abstract
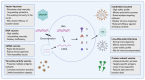
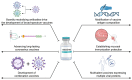
The continuous evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the emergence of variants of concern (VOCs) underscore the critical role of vaccination in pandemic control. These mutations not only enhance viral infectivity but also facilitate immune evasion and diminish vaccine efficacy, necessitating ongoing surveillance and vaccine adaptation. Current SARS-CoV-2 vaccines, including inactivated, live-attenuated, viral vector, protein subunit, virus-like particle, and nucleic acid vaccines, face challenges due to the immune evasion strategies of emerging variants. Moreover, other sarbecoviruses, such as SARS-CoV-1 and SARS-related coronaviruses (SARSr-CoVs) pose a potential risk for future outbreaks. Thus, developing vaccines capable of countering emerging SARS-CoV-2 variants and providing broad protection against multiple sarbecoviruses is imperative. Several innovative vaccine platforms are being investigated to elicit broad-spectrum neutralizing antibody responses, offering protection against both current SARS-CoV-2 variants and other sarbecoviruses. This review presents an updated overview of the key target antigens and therapeutic strategies employed in current SARS-CoV-2 vaccines. Additionally, we summarize ongoing approaches for the development of vaccines targeting infectious sarbecoviruses.
Keywords: broadly neutralizing antibodies; coronavirus; immune evasion; sarbecovirus; vaccines.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
A critical residue in a conserved RBD epitope determines neutralization breadth of pan-sarbecovirus antibodies with recurring YYDRxxG motifs.mBio. 2025 Jul 31:e0060625. doi: 10.1128/mbio.00606-25. Online ahead of print. mBio. 2025. PMID: 40742150
-
Efficacy and safety of COVID-19 vaccines.Cochrane Database Syst Rev. 2022 Dec 7;12(12):CD015477. doi: 10.1002/14651858.CD015477. Cochrane Database Syst Rev. 2022. PMID: 36473651 Free PMC article.
-
Workplace interventions to reduce the risk of SARS-CoV-2 infection outside of healthcare settings.Cochrane Database Syst Rev. 2022 May 6;5(5):CD015112. doi: 10.1002/14651858.CD015112.pub2. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 Apr 10;4:CD015112. doi: 10.1002/14651858.CD015112.pub3. PMID: 35514111 Free PMC article. Updated.
-
Detection of SARS-CoV-2-Specific Antibodies in Human Breast Milk and Their Neutralizing Capacity after COVID-19 Vaccination: A Systematic Review.Int J Mol Sci. 2023 Feb 3;24(3):2957. doi: 10.3390/ijms24032957. Int J Mol Sci. 2023. PMID: 36769279 Free PMC article.
-
The pan-variant potential of light: 425 nm light inactivates SARS-CoV-2 variants of concern and non-cytotoxic doses reduce viral titers in human airway epithelial cells.mSphere. 2025 Jun 25;10(6):e0023025. doi: 10.1128/msphere.00230-25. Epub 2025 May 28. mSphere. 2025. PMID: 40434113 Free PMC article.
References
Publication types
Grants and funding
- 2023YFC2605400, 2023YFC3404000/National Key Research and Development Program of China
- 32300121, 32270142/National Natural Science Foundation of China
- 92169212/Major Research Plan of the National Natural Science Foundation of China
- 23PJD007/Shanghai Pujiang Programme
- 22QA1408800/Shanghai Rising-Star Program
LinkOut - more resources
Full Text Sources
Miscellaneous

