Dhdds T206A and Dhdds K42E knock-in mouse models of retinitis pigmentosa 59 are phenotypically similar
- PMID: 40574710
- PMCID: PMC12352288
- DOI: 10.1242/dmm.052243
Dhdds T206A and Dhdds K42E knock-in mouse models of retinitis pigmentosa 59 are phenotypically similar
Abstract
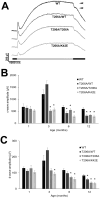
Dehydrodolichyl diphosphate synthase complex subunit (DHDDS) is required for protein glycosylation in eukaryotes, and variants. Surprisingly, three variant alleles (K42E/K42E, T206A/K42E and R98W/K42E) have been reported to cause retinitis pigmentosa 59 (RP59). Because T206A only has been reported to occur heterozygously with K42E, we generated homozygous and hererozygous mutants - i.e. T206A/T206A and T206A/K42E, respectively - in mice to assess the effect of the T206A allele. By postnatal age of 12 month (PN 12-mo), T206A/T206A and T206A/K42E mice exhibited reduction of inner nuclear layer thickness as observed in K42E/K42E mice. Electroretinography (ERG) revealed a reduction in b-waves, but spared reduction in a-wave amplitudes. By PN 3-mo, ERG c- and d-waves were significantly attenuated in all phenotypes. Consistent with a reduction in inner nuclear layer thickness as seen by using optical coherence tomography (OCT), cell loss observed by histology, as well as bipolar and amacrine cell densities were reduced in all Dhdds mutant phenotypes compared to those of PN 8-12 mo age-matched controls. These results indicated that the DHDDS T206A allele, like the K42E allele, causes retinal disease, probably through a common pathobiological mechanism. We propose that the physiological basis of retinal dysfunction in RP59 involves defective photoreceptor to bipolar cell synaptic transmission with concomitant bipolar/amacrine cell degeneration.
Keywords: Dolichol; Glycosylation; Inherited retinal degeneration; Mouse models; RP59; Retinitis pigmentosa.
© 2025. Published by The Company of Biologists.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures







Update of
-
Modeling Retinitis Pigmentosa 59: Dhdds T206A and Dhdds K42E knock-in mutant mice are phenotypically similar.bioRxiv [Preprint]. 2025 Jan 15:2025.01.13.632845. doi: 10.1101/2025.01.13.632845. bioRxiv. 2025. Update in: Dis Model Mech. 2025 Jul 1;18(7):dmm052243. doi: 10.1242/dmm.052243. PMID: 39868263 Free PMC article. Updated. Preprint.
Similar articles
-
Modeling Retinitis Pigmentosa 59: Dhdds T206A and Dhdds K42E knock-in mutant mice are phenotypically similar.bioRxiv [Preprint]. 2025 Jan 15:2025.01.13.632845. doi: 10.1101/2025.01.13.632845. bioRxiv. 2025. Update in: Dis Model Mech. 2025 Jul 1;18(7):dmm052243. doi: 10.1242/dmm.052243. PMID: 39868263 Free PMC article. Updated. Preprint.
-
A Dhdds K42E knock-in RP59 mouse model shows inner retina pathology and defective synaptic transmission.Cell Death Dis. 2023 Jul 13;14(7):420. doi: 10.1038/s41419-023-05936-4. Cell Death Dis. 2023. PMID: 37443173 Free PMC article.
-
Lack of Overt Retinal Degeneration in a K42E Dhdds Knock-In Mouse Model of RP59.Cells. 2020 Apr 7;9(4):896. doi: 10.3390/cells9040896. Cells. 2020. PMID: 32272552 Free PMC article.
-
Vitamin A and fish oils for retinitis pigmentosa.Cochrane Database Syst Rev. 2013 Dec 19;2013(12):CD008428. doi: 10.1002/14651858.CD008428.pub2. Cochrane Database Syst Rev. 2013. Update in: Cochrane Database Syst Rev. 2020 Jun 18;6(6):CD008428. doi: 10.1002/14651858.CD008428.pub3. PMID: 24357340 Free PMC article. Updated.
-
The Road towards Gene Therapy for X-Linked Juvenile Retinoschisis: A Systematic Review of Preclinical Gene Therapy in Cell-Based and Rodent Models of XLRS.Int J Mol Sci. 2024 Jan 19;25(2):1267. doi: 10.3390/ijms25021267. Int J Mol Sci. 2024. PMID: 38279267 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- UL1 TR003096/TR/NCATS NIH HHS/United States
- F31 EY032764/EY/NEI NIH HHS/United States
- University of Alabama at Birmingham
- I K6 BX005787/Biomedical Laboratory Research and Development, VA Office of Research and Development
- P60 DK079626/DK/NIDDK NIH HHS/United States
- IK6 BX005787/BX/BLRD VA/United States
- UL1TR003096/TR/NCATS NIH HHS/United States
- P30 DK074038/DK/NIDDK NIH HHS/United States
- UL1TR003096/National Center for Advancing Translational Sciences (NCATS)
- P30 CA13148/CA/NCI NIH HHS/United States
- R01 EY029341/EY/NEI NIH HHS/United States
- VA Western New York Healthcare System
- P30 DK079626/DK/NIDDK NIH HHS/United States
- P30 CA013148/CA/NCI NIH HHS/United States
- U.S. Department of Health and Human Services
- P30 AR048311/AR/NIAMS NIH HHS/United States
- P30 EY003039/EY/NEI NIH HHS/United States
- U.S. Department of Veterans Affairs
- DK05336/DK/NIDDK NIH HHS/United States
- University of Alabama (UAB)
- National Institutes of Health (NIH)
- EY029341/EY/NEI NIH HHS/United States
- University at Buffalo
LinkOut - more resources
Full Text Sources

