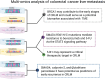
Multi-omics in colorectal cancer liver metastasis: applications and research advances
- PMID: 40574729
- PMCID: PMC12240200
- DOI: 10.20892/j.issn.2095-3941.2025.0066
Multi-omics in colorectal cancer liver metastasis: applications and research advances
Abstract
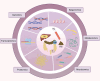
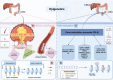
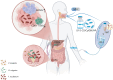
Colorectal cancer (CRC) is a common malignant tumor with a high mortality rate worldwide. Advanced CRC often leads to liver metastasis, which has a poor prognosis, highlighting the need to investigate the underlying mechanisms. Omics, encompassing genomics, epigenomics, transcriptomics, proteomics, metabolomics, and microbiomics, enables comprehensive molecular analysis of cells and tissues. Tumor-omics research has advanced rapidly, with growing attention on CRC-related omics. However, systematic reviews on omics research specific to colorectal cancer liver metastasis (CRLM) are limited. This review summarizes the current status and progress of multi-omics research on CRLM and discusses the application of multi-omics technologies in basic research and the significant clinical implications.
Keywords: Colorectal cancer liver metastasis; epigenomics; genomics; metabolomics; microbiomics; proteomics; transcriptomics.
Copyright © 2025 The Authors.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Figures
Similar articles
-
Multi-omics approaches: transforming the landscape of natural product isolation.Funct Integr Genomics. 2025 Jun 19;25(1):132. doi: 10.1007/s10142-025-01645-7. Funct Integr Genomics. 2025. PMID: 40537580 Review.
-
Spatial multi-omics reveals the potential involvement of SPP1+ fibroblasts in determining metabolic heterogeneity and promoting metastatic growth of colorectal cancer liver metastasis.Mol Ther. 2025 Aug 6;33(8):3680-3700. doi: 10.1016/j.ymthe.2025.05.004. Epub 2025 May 7. Mol Ther. 2025. PMID: 40340245
-
[Advances in Multi-omics Analysis of Cervical Cancer].Sichuan Da Xue Xue Bao Yi Xue Ban. 2025 Mar 20;56(2):571-576. doi: 10.12182/20250360403. Sichuan Da Xue Xue Bao Yi Xue Ban. 2025. PMID: 40599295 Free PMC article. Review. Chinese.
-
Towards a Multi-omics Understanding of Anaphylaxis: Insights into Pathogenesis and Biomarker Identification.Clin Rev Allergy Immunol. 2025 Jun 30;68(1):61. doi: 10.1007/s12016-025-09069-8. Clin Rev Allergy Immunol. 2025. PMID: 40588688 Free PMC article. Review.
-
Multi-omics Combined with Machine Learning Facilitating the Diagnosis of Gastric Cancer.Curr Med Chem. 2024;31(40):6692-6712. doi: 10.2174/0109298673284520240112055108. Curr Med Chem. 2024. PMID: 38351697 Review.
Cited by
-
The dark matter in cancer immunology: beyond the visible- unveiling multiomics pathways to breakthrough therapies.J Transl Med. 2025 Jul 22;23(1):808. doi: 10.1186/s12967-025-06839-y. J Transl Med. 2025. PMID: 40696449 Free PMC article. Review.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Zhao J, Yang J. Multi-omics research for precision medicine. Beijing: Science Press; 2021.
-
- Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, Lee WS, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
