The genome sequence of the greater argentine, Argentina silus (Ascanius, 1775)
- PMID: 40574744
- PMCID: PMC12198749
- DOI: 10.12688/wellcomeopenres.23691.1
The genome sequence of the greater argentine, Argentina silus (Ascanius, 1775)
Abstract
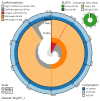
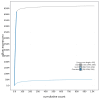
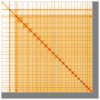
We present a genome assembly from a male specimen of Argentina silus (the greater argentine; Chordata; Actinopteri; Argentiniformes; Argentinidae). The genome sequence has a total length of 670.70 megabases. Most of the assembly is scaffolded into 24 chromosomal pseudomolecules, including the X and Y sex chromosomes. The mitochondrial genome has also been assembled and is 16.64 kilobases in length. Gene annotation of this assembly on Ensembl identified 19,422 protein-coding genes.
Keywords: Argentina silus; Argentiniformes; chromosomal; genome sequence; greater argentine.
Copyright: © 2025 Vang HBM et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
The genome sequence of Atlantic Bluefin Tuna, Thunnus thynnus (Linnaeus, 1758).Wellcome Open Res. 2025 Mar 27;10:163. doi: 10.12688/wellcomeopenres.23971.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40556665 Free PMC article.
-
The genome sequence of a soldier fly, Nemotelus pantherinus (Linnaeus, 1758).Wellcome Open Res. 2025 Jun 2;10:303. doi: 10.12688/wellcomeopenres.24309.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40548330 Free PMC article.
-
The genome sequence of the Asparagus Beetle, Crioceris asparagi (Linnaeus, 1758).Wellcome Open Res. 2025 Jan 15;10:18. doi: 10.12688/wellcomeopenres.23460.1. eCollection 2025. Wellcome Open Res. 2025. PMID: 40528997 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
-
Single-incision sling operations for urinary incontinence in women.Cochrane Database Syst Rev. 2017 Jul 26;7(7):CD008709. doi: 10.1002/14651858.CD008709.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Oct 27;10:CD008709. doi: 10.1002/14651858.CD008709.pub4. PMID: 28746980 Free PMC article. Updated.
References
-
- Árnason E: A report on the mitochondrial DNA variation of the great silver smelt. Argentina silus,2017.
LinkOut - more resources
Full Text Sources

