Emergence and transmission dynamics of the FY.4 Omicron variant in Kenya
- PMID: 40574750
- PMCID: PMC12202047
- DOI: 10.1093/ve/veaf035
Emergence and transmission dynamics of the FY.4 Omicron variant in Kenya
Abstract
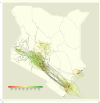
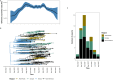
The recombinant FY.4 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant was first reported in Kenya in March 2023 and was the dominant circulating variant between April and July 2023. The variant was characterized by two important mutations: Y451H in the receptor-binding domain of the spike protein and P42L in open reading frame 3a. Using phylogenetics and phylodynamic approaches, we investigated the emergence and spread of FY.4 in Kenya and the rest of the world. Our findings suggest FY.4 circulated early in Kenya before its export to North America and Europe. Early circulation of FY.4 in Kenya was predominantly observed in the coastal part of the country, and the estimated time to the most recent common ancestor suggests FY.4 circulated as early as December 2022. The collected genomic and epidemiological data show that the FY.4 variant led to a large local outbreak in Kenya and resulted in localized outbreaks in Europe, North America, and Asia-Pacific. These findings underscore the importance of sustained genomic surveillance, especially in under-sampled regions, in deepening our understanding of the evolution and spread of SARS-CoV-2 variants.
Keywords: FY.4; Kenya; Omicron; phylogenetics.
© The Author(s) 2025. Published by Oxford University Press.
Figures


Similar articles
-
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya.mSphere. 2025 Jun 25;10(6):e0002725. doi: 10.1128/msphere.00027-25. Epub 2025 May 29. mSphere. 2025. PMID: 40439429 Free PMC article.
-
New SARS-CoV-2 Omicron Variant with Spike Protein Mutation Y451H, Kilifi, Kenya, March-May 2023.Emerg Infect Dis. 2023 Nov;29(11):2376-2379. doi: 10.3201/eid2911.230894. Epub 2023 Sep 14. Emerg Infect Dis. 2023. PMID: 37708843 Free PMC article.
-
The pan-variant potential of light: 425 nm light inactivates SARS-CoV-2 variants of concern and non-cytotoxic doses reduce viral titers in human airway epithelial cells.mSphere. 2025 Jun 25;10(6):e0023025. doi: 10.1128/msphere.00230-25. Epub 2025 May 28. mSphere. 2025. PMID: 40434113 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
References
-
- Center for Disease Control and Prevention . SARS-CoV-2 Variant Classifications and Definitions, 2021. https://stacks.cdc.gov/view/cdc/105817
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

