Riboflavin transporter: evidence of a role as entry receptor for chimpanzee endogenous retrovirus
- PMID: 40574751
- PMCID: PMC12202039
- DOI: 10.1093/ve/veaf031
Riboflavin transporter: evidence of a role as entry receptor for chimpanzee endogenous retrovirus
Abstract
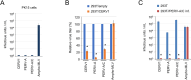
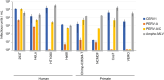
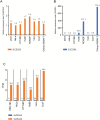
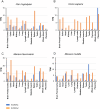
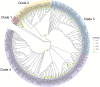
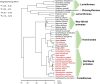
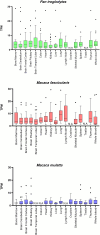
Endogenous retroviruses (ERVs) are remnants of ancestral viral infections in germ cells that constitute a substantial proportion of the mammalian genome and are assumed to provide molecular fossil records of ancient infections. Analysis of these sequences may reveal the mechanisms of virus-host co-evolution, viral endogenization, and extinction. Chimpanzee endogenous retrovirus 1 (CERV1), a gamma retrovirus, is estimated to have circulated within primates for ~10 million years, although it is now apparently extinct. In this study, we aimed to gain an understanding of how the extinct CERV1 was transmitted and endogenized. On the basis of the identification of CERV1 fossils in the primate genome and using the expression-cloning method with the human cDNA library, we found that riboflavin transporter human SLC52A2 served as a receptor for CERV1 entry. The ectopic expression of human and chimpanzee SLC52A2 and its related SLC52A1 in heterogenic cells confers susceptibility to infection by CERV1 and porcine endogenous retrovirus (PERV). Virus interference experiments have shown that CERV1 inhibits infection by PERV and vice versa. This finding indicates that CERV1 and PERV belong to the same virus interference group. CERV1 shows infection in a wide range of human and primate cells. Notably, CERV1 infection is observed in human cell lines that express human SLC52A2 abundantly but hardly express human SLC52A1. Although CERV1 has been established to be present at high copy numbers in the great apes (Pan troglodytes, Pan paniscus, and Gorilla gorilla) and 15 Old World monkey species of the Cercopithecinae and Colobinae subfamilies, it is absent in humans and orangutans. CERV1 gene expression is observed in primates, including chimpanzees, suggesting that CERV1 has co-evolved with its hosts. Our results suggest that ERVs may have conferred resistance to viral infections in a convergent evolutionary manner. These findings are significant not only for advancing the field of paleovirology but also in terms of gaining an understanding of the potential risks of viral infection with respect to xenotransplantation, such as that from pigs to humans.
Keywords: CERV1; endogenous retrovirus; paleovirology; riboflavin transporter.
© The Author(s) 2025. Published by Oxford University Press.
Figures

Similar articles
-
NIH Consensus Statement on Management of Hepatitis C: 2002.NIH Consens State Sci Statements. 2002 Jun 10-12;19(3):1-46. NIH Consens State Sci Statements. 2002. PMID: 14768714
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Single-incision sling operations for urinary incontinence in women.Cochrane Database Syst Rev. 2017 Jul 26;7(7):CD008709. doi: 10.1002/14651858.CD008709.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Oct 27;10:CD008709. doi: 10.1002/14651858.CD008709.pub4. PMID: 28746980 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
References
LinkOut - more resources
Full Text Sources

