Multi-omics-based subtyping of melanoma suggests distinct immune and targeted therapy strategies
- PMID: 40574832
- PMCID: PMC12197953
- DOI: 10.3389/fimmu.2025.1601243
Multi-omics-based subtyping of melanoma suggests distinct immune and targeted therapy strategies
Abstract
Background: Melanoma is a highly heterogeneous malignancy with diverse molecular and clinical behaviors. A precise molecular classification is critical for improving prognostic assessment and guiding personalized therapy.
Methods: We performed an integrative multi-omics analysis of skin cutaneous melanoma using data from The Cancer Genome Atlas (TCGA) and validated our findings in independent cohorts. Multi-layered data, including transcriptomic, genomic, epigenetic, and immune landscape profiles, were analyzed using unsupervised clustering and machine learning approaches to define molecular subtypes. Functional assays and in silico drug screening were employed to explore subtype-specific vulnerabilities.
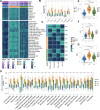
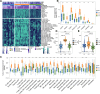
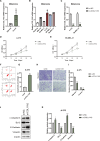
Results: Three robust molecular subtypes (CS1, CS2, CS3) were identified, each with distinct genomic alterations, tumor microenvironment characteristics, and clinical outcomes. The CS2 subtype was immunologically "hot," characterized by high tumor mutational burden (TMB), elevated neoantigen load, strong immune infiltration, and activated IFN-γ signaling. CS2 tumors showed significant enrichment of immune checkpoint gene expression and were associated with favorable response to anti-PD-1 therapy in external validation cohorts. In contrast, CS1 and CS3 were immunologically "cold" with immune exclusion, high chromosomal instability, and activation of oncogenic pathways linked to immune evasion. Transcriptomic drug sensitivity modeling suggested that CS1 and CS3 may benefit from HSP90 or MEK inhibitors. Moreover, COL11A2 was identified as a subtype-enriched oncogenic driver predominantly expressed in CS1/CS3, and its silencing impaired tumor cell proliferation, invasion, and epithelial-mesenchymal transition (EMT) features.
Conclusions: This study presents a refined multi-omics classification of melanoma that reveals biologically and clinically distinct subtypes with divergent immune and therapeutic profiles. It offers a framework for subtype-specific treatment strategies, and identifies COL11A2 as a potential target in immune-cold melanomas.
Keywords: immune checkpoint therapy; in-silico drug screening; melanoma; molecular subtypes; multi-omics integration.
Copyright © 2025 Li, Lin, Wang, Zhou, Feng and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Identification and validation of a KRAS-macrophage-associated gene signature as prognostic biomarkers and potential therapeutic targets in melanoma.Front Immunol. 2025 Jun 18;16:1566432. doi: 10.3389/fimmu.2025.1566432. eCollection 2025. Front Immunol. 2025. PMID: 40607411 Free PMC article.
-
Comprehensive multi-omics and machine learning framework for glioma subtyping and precision therapeutics.Sci Rep. 2025 Jul 10;15(1):24874. doi: 10.1038/s41598-025-09742-0. Sci Rep. 2025. PMID: 40640380 Free PMC article.
-
Construction and validation of a lipid metabolism-related genes prognostic signature for skin cutaneous melanoma.Biochem Biophys Res Commun. 2025 Aug 15;775:152115. doi: 10.1016/j.bbrc.2025.152115. Epub 2025 May 29. Biochem Biophys Res Commun. 2025. PMID: 40460484
-
Neoadjuvant treatment for stage III and IV cutaneous melanoma.Cochrane Database Syst Rev. 2023 Jan 17;1(1):CD012974. doi: 10.1002/14651858.CD012974.pub2. Cochrane Database Syst Rev. 2023. PMID: 36648215 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

