Single cell dissection reveals SFRP2+ fibroblasts amplifying inflammatory responses in oral lichen planus
- PMID: 40574847
- PMCID: PMC12197934
- DOI: 10.3389/fimmu.2025.1553963
Single cell dissection reveals SFRP2+ fibroblasts amplifying inflammatory responses in oral lichen planus
Abstract
Objectives: Oral lichen planus (OLP) is a chronic inflammatory mucosal disease with an incompletely understood pathogenesis. This study aimed to investigate the role of disease-specific fibroblasts in OLP.
Methods: We performed single-cell RNA sequencing on buccal mucosa of 4 OLP patients and one healthy control. Additionally, mRNA expression and immunofluorescence staining were analyzed in primary fibroblasts from 51 OLP patients and 24 healthy individuals. The spatial cellular interactions were assessed using multiplex immunofluorescences in OLP tissues.
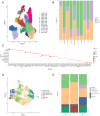
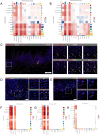
Results: Using single-cell RNA sequencing, we identified SFRP2+ fibroblasts as the origin of inflammatory fibroblasts in OLP. A subset of SFRP2+ fibroblasts specifically expressed Wnt5a and was implicated in antigen processing and presentation pathway in OLP. Furthermore, SFRP2+Wnt5a+ fibroblasts amplified and maintained the local immune inflammation by interacting with CD8+ T cells and epithelial cells. Compared to the healthy control group, upregulated expressions of pro-inflammatory molecules (CXCL12, CXCL14) and antigen presenting associated molecules (HLA-A, HLA-B, HLA-C and ERAP2) were displayed in OLP group at mRNA level. Colocalization of SFRP2 and Wnt5a was displayed in the primary cultured fibroblasts of OLP in vitro. Besides, SFRP2+ Wnt5a+ fibroblasts were located around CD8+ T cells in the superficial layer of the lymphocyte infiltration zone.
Conclusions: Our findings reveal the heterogeneity and pathogenic mechanisms of fibroblasts in OLP, providing new insights into the cellular drivers of chronic inflammation in OLP.
Keywords: SFRP2; antigen processing and presenting; fibroblasts; oral lichen planus; single-cell sequencing.
Copyright © 2025 Cheng, Liu, Zhu, Yang, Geng and Fan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Single-Cell RNA Sequencing Reveals Functional Exhaustion of T Cells in Oral Lichen Planus.Mol Oral Microbiol. 2025 Aug;40(4):147-157. doi: 10.1111/omi.12495. Epub 2025 May 14. Mol Oral Microbiol. 2025. PMID: 40366262
-
DnaK of Parvimonas micra extracellular vesicles interacts with the host fibroblasts BAG3-IKK-γ axis to accelerate TNF-α secretion in oral lichen planus.Microbiome. 2025 Jul 14;13(1):164. doi: 10.1186/s40168-025-02151-5. Microbiome. 2025. PMID: 40660385 Free PMC article.
-
B Cells Infiltration Potentially Responded Better to Systemic Corticoids in Oral Lichen Planus and Oral Lichenoid Lesions.Inflammation. 2025 Jun;48(3):1235-1244. doi: 10.1007/s10753-024-02112-4. Epub 2024 Aug 9. Inflammation. 2025. PMID: 39117788
-
A Systematic Review of Interleukin-17 in Oral Lichen Planus: From Etiopathogenesis to Treatment.Clin Med Res. 2023 Dec;21(4):201-215. doi: 10.3121/cmr.2023.1822. Clin Med Res. 2023. PMID: 38296640 Free PMC article.
-
Interventions for treating oral lichen planus.Cochrane Database Syst Rev. 2011 Jul 6;(7):CD001168. doi: 10.1002/14651858.CD001168.pub2. Cochrane Database Syst Rev. 2011. Update in: Cochrane Database Syst Rev. 2020 Feb 28;2:CD001168. doi: 10.1002/14651858.CD001168.pub3. PMID: 21735381 Updated.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

