Broad PFAS Binding with Fatty Acid Binding Protein 4 Is Enabled by Variable Binding Modes
- PMID: 40575325
- PMCID: PMC12188406
- DOI: 10.1021/jacsau.5c00504
Broad PFAS Binding with Fatty Acid Binding Protein 4 Is Enabled by Variable Binding Modes
Abstract
Per- and polyfluoroalkyl substances (PFAS) are ubiquitous pollutants that bioaccumulate in wildlife and humans, yet the molecular basis of their protein interactions remains poorly understood. Here, we show that human adipocyte fatty acid-binding protein 4 (FABP4) can bind a diverse array of PFAS, including next-generation replacements for legacy chemicals and longer-chain perfluorocarboxylic acids. Shorter-chain PFAS, although weaker binders, still displayed measurable affinities, surpassing those of their nonfluorinated analogs. We determined crystal structures of FABP4 bound to perfluorooctanoic acid (PFOA), perfluorodecanoic acid (PFDA), and perfluorohexadecanoic acid (PFHxDA), revealing three distinct binding modes. Notably, PFOA binds in two separate sites, and two distinct conformations define single-ligand binding of PFDA and PFHxDA. These arrangements enhance hydrophobic interactions within the binding cavity and likely explain the low micromolar dissociation constants observed in fluorescence competition assays. Our findings underscore the critical roles of chain length, headgroup functionality, and protein conformation in PFAS-FABP4 interactions. Given the emerging implications of the role of FABP4 in endocrine function, even subtle PFAS-induced perturbations could affect metabolic regulation and disease risk. Overall, this work highlights the value of direct structural and biochemical insights into PFAS-FABP4 interactions and paves the way for future research on PFAS transport and toxicological outcomes.
Keywords: FABP4; Fatty acid binding protein 4; PFAS; X-ray crystallography; fluorinated ligands; per- and polyfluoroalkyl substances; protein structure.
© 2025 The Authors. Published by American Chemical Society.
Figures

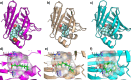

Update of
-
Broad PFAS binding with fatty acid binding protein 4 is enabled by variable binding modes.bioRxiv [Preprint]. 2025 Jan 14:2025.01.10.632451. doi: 10.1101/2025.01.10.632451. bioRxiv. 2025. Update in: JACS Au. 2025 Jun 02;5(6):2469-2474. doi: 10.1021/jacsau.5c00504. PMID: 40196552 Free PMC article. Updated. Preprint.
References
-
- Abunada Z., Alazaiza M. Y. D., Bashir M. J. K.. An Overview of Per- and Polyfluoroalkyl Substances (PFAS) in the Environment: Source, Fate, Risk and Regulations. Water (Basel) 2020;12(12):3590. doi: 10.3390/w12123590. - DOI
-
- De Silva A. O., Armitage J. M., Bruton T. A., Dassuncao C., Heiger-Bernays W., Hu X. C., Kärrman A., Kelly B., Ng C., Robuck A., Sun M., Webster T. F., Sunderland E. M.. PFAS Exposure Pathways for Humans and Wildlife: A Synthesis of Current Knowledge and Key Gaps in Understanding. Environ. Toxicol. Chem. 2020;40(3):631–657. doi: 10.1002/etc.4935. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
