The significance of metagenomic next-generation sequencing and targeted next-generation sequencing in the diagnostic evaluation of patients with suspected pulmonary infections
- PMID: 40575480
- PMCID: PMC12198251
- DOI: 10.3389/fcimb.2025.1552236
The significance of metagenomic next-generation sequencing and targeted next-generation sequencing in the diagnostic evaluation of patients with suspected pulmonary infections
Abstract
Objective: To investigate the diagnostic value of metagenomic next-generation sequencing (mNGS) and targeted next-generation sequencing (tNGS) in identifying pathogens in patients with pulmonary infections.
Methods: A retrospective analysis was conducted on 155 patients with suspected lung infections who underwent alveolar lavage and were admitted to the Department of Respiratory and Critical Care Medicine at Baodi Hospital, Tianjin Medical University, from July 2023 to December 2023. The bronchoalveolar lavage fluid (BALF) samples obtained were subjected to mNGS, tNGS and culture methods to compare their diagnostic efficacy in identifying lung infection pathogens.
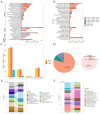
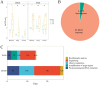
Results: The results indicated that both tNGS and mNGS methods exhibit comparable detection efficiencies in identifying pathogens in patients with pulmonary infections, significantly outperforming BALF culture approach. In terms of diagnostic accuracy, tNGS exhibited a higher sensitivity than mNGS, with rates of 96.1% and 75.7% respectively (P>0.05). However, the specificity of tNGS was slightly lower than that of mNGS, with rates of 59.1% and 68.2% respectively (P>0.05). It is noteworthy that this difference in specificity was not statistically significant.
Conclusion: tNGS exhibits a diagnostic efficacy comparable to mNGS, particularly in its sensitivity for identifying lung infections, as evidenced by expert insights and clinical applications. Furthermore, tNGS offers advantages in convenience, time efficiency, and cost-effectiveness, hinting at its potential to serve as an alternative to mNGS in clinical settings.
Keywords: bronchoalveolar lavage fluid; mNGS; pathogens; pulmonary infection; tNGS.
Copyright © 2025 Tang, Tang, Zhang, Lin and Shan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Comparison of the diagnostic capabilities of tNGS and mNGS for pathogens causing lower respiratory tract infections: a prospective observational study.Front Cell Infect Microbiol. 2025 Jun 10;15:1578939. doi: 10.3389/fcimb.2025.1578939. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40557317 Free PMC article.
-
Application of targeted next-generation sequencing for pathogens diagnosis and drug resistance prediction in bronchoalveolar lavage fluid of pulmonary infections.Front Cell Infect Microbiol. 2025 Jun 9;15:1590881. doi: 10.3389/fcimb.2025.1590881. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40552119 Free PMC article.
-
Performance of broad-spectrum targeted next-generation sequencing in lower respiratory tract infections in ICU patients: a prospective observational study.Crit Care. 2025 Jun 4;29(1):226. doi: 10.1186/s13054-025-05470-z. Crit Care. 2025. PMID: 40468429 Free PMC article.
-
Diagnostic accuracy of metagenomic next-generation sequencing in pulmonary tuberculosis: a systematic review and meta-analysis.Syst Rev. 2024 Dec 27;13(1):317. doi: 10.1186/s13643-024-02733-8. Syst Rev. 2024. PMID: 39731100 Free PMC article.
-
The diagnostic efficacy of metagenomic next-generation sequencing (mNGS) in pathogen identification of pediatric pneumonia using bronchoalveolar lavage fluid (BALF): A systematic review and meta-analysis.Microb Pathog. 2025 Jun;203:107492. doi: 10.1016/j.micpath.2025.107492. Epub 2025 Mar 18. Microb Pathog. 2025. PMID: 40113108
Cited by
-
Recurrent Cough in a Pediatric Patient From China: A Case Report of Bordetella pertussis Infection With Genomic Insights.Cureus. 2025 Jun 25;17(6):e86760. doi: 10.7759/cureus.86760. eCollection 2025 Jun. Cureus. 2025. PMID: 40718202 Free PMC article.
References
-
- Cassini A., Högberg L. D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G. S., et al. (2019). Attributab le deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66. doi: 10.1016/S1473-3099(18)30605-4 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

