Genomic evolution and dissemination of non-conjugative virulence plasmid of ST65 carbapenem-resistant and hypervirulent Klebsiella pneumoniae strains in a Chinese hospital
- PMID: 40575489
- PMCID: PMC12198223
- DOI: 10.3389/fcimb.2025.1548300
Genomic evolution and dissemination of non-conjugative virulence plasmid of ST65 carbapenem-resistant and hypervirulent Klebsiella pneumoniae strains in a Chinese hospital
Abstract
Background: The global rise in infections caused by hypervirulent and carbapenem-resistant K. pneumoniae (CR-hvKp) represents a growing public health threat. This study investigates ST65 CR-hvKp strains, with a focus on their genomic attributes and the mechanisms underlying the transmission of non-conjugative virulence plasmids.
Methods: Two clinical K2-ST65 CR-hvKp isolates (P6 and P10) were identified. Plasmid conjugation experiments were performed to assess the dissemination of the virulence plasmid. Antimicrobial susceptibility testing and virulence assays, including serum resistance, siderophore production, and G. mellonella larvae infection models, were used to characterize resistance and virulence phenotypes. Comprehensive bioinformatic analyses were performed to explore genetic evolution.
Results: Genomic analyses showed that both P6 and P10 carry a non-conjugative virulence plasmid, a conjugative untyped KPC plasmid, and a conjugative IncM2 plasmid. These isolates displayed broad-spectrum anti-microbial resistance and multiple virulence phenotypes, although they failed to sustain both hypervirulence and carbapenem resistance over time. The IncM2 plasmid was shown to be essential for the transfer of non-conjugative virulence plasmid. Mechanistic studies highlighted IS26-mediated plasmid fusion and the role of IncM2 plasmids in mobilizing non-conjugative virulence plasmids. The resulting transconjugants exhibited multidrug resistance, enhanced capsule production, and increased siderophore production.
Conclusions: This study provides new insights into the genomic dynamics of ST65-CR-hvKp strains and uncovers key mechanisms, such as IS26-mediated plasmid fusion and IncM2-mediated mobilization, which facilitate the dissemination of non-conjugative virulence plasmids. Understanding these mechanisms is crucial for developing effective strategies to manage and prevent the spread of these clinically challenging strains.
Keywords: Klebsiella pneumoniae; ST65; carbapenem resistance; hypervirulence; plasmid transmission.
Copyright © 2025 Tian, Zhao, Liu, Lu and Dong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

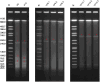
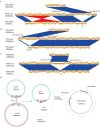
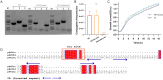

References
-
- Al Bshabshe A., Al-Hakami A., Alshehri B., Al-Shahrani K. A., Alshehri A. A., Al Shahrani M. B., et al. (2020). Rising klebsiella pneumoniae infections and its expanding drug resistance in the intensive care unit of a tertiary healthcare hospital, Saudi Arabia. Cureus 12, e10060. doi: 10.7759/cureus.10060 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

