Alpha to JN.1 variants: SARS-CoV-2 genomic analysis unfolding its various lineages/sublineages evolved in Chhattisgarh, India from 2020 to 2024
- PMID: 40575649
- PMCID: PMC12188907
- DOI: 10.5501/wjv.v14.i2.100001
Alpha to JN.1 variants: SARS-CoV-2 genomic analysis unfolding its various lineages/sublineages evolved in Chhattisgarh, India from 2020 to 2024
Abstract
Background: The evolutionary mutational changes of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) since its emergence in Chhattisgarh, India in 2020 have warranted the need for the characterization of every lineage/sublineage that has evolved until February 2024.
Aim: To unravel the evolutionary pathway of SARS-CoV-2 in Chhattisgarh from 2020 to February 2024.
Methods: A total of 635 coronavirus disease 2019 cases obtained between 2020 and February 2024 were investigated by whole genome sequencing.
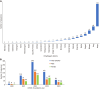
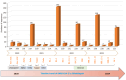
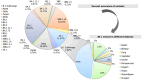
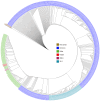
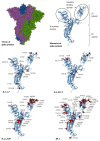
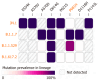
Results: Whole genome sequencing analysis identified the evolution of SARS-CoV-2 into seventeen lineages from 2020 to 2024. SARS-CoV-2 initially emerged in Chhattisgarh in its Alpha (B.1.1.7) variant in 2020. Thereafter, it continuously underwent periodical mutational changes in the spike gene to further differentiate into various lineages/sublineages, viz., Kappa, Delta, BA.1, and BA.2 in 2021; the Omicron lineage (BA.5, BA.2.12.1, BA.2.75, BQ.1, and XBB) in 2022; the new Omicron lineage (XBB.1.5, XBB.1.16, XBB.1.9.1, and XBB.2.3) in 2023; and finally to JN.1 in January and February 2024. The predominant lineages over these 4 years were BA.1.1.7 (Alpha) in 2020, B.1.617.2 (Delta) in the period between 2021 and mid-2022, B.1.1.529 (Omicron) in late 2022 to 2023, and Omicron-JN.1 in early 2024. The presently circulating JN.1 lineage was observed harboring exclusive predominant mutations of E4554K, A570V, P621A, and P1143 L with 99% prevalence.
Conclusion: SARS-CoV-2 from 2020 to 2024 has evolved into 17 lineages/sublineages in Chhattisgarh. The presently circulating JN.1 harbored 40 mutations, especially E554K, A570V, P621S, and P1143 L, capacitating the virus with features of host cell entry, stability, replication, rapid transmissibility, and crucial immune evasion. Therefore, earlier immunity from either vaccination or prior infection may not protect against the current lineage and increases the possibility of future outbreaks. Thus, the periodical genomic surveillance of SARS-CoV-2 is essential for the genomic blueprint of the circulating virus, which may help in updating the vaccine strain and various basic research for developing appropriate therapeutics and diagnostics.
Keywords: 2020 to 2024; Genomic surveillance; Genomic variants; JN.1 variant; SARS-CoV-2.
©The Author(s) 2025. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors have no conflicts of interest regarding the publication of this article.
Figures
Similar articles
-
An mRNA vaccine encoding the SARS-CoV-2 Omicron XBB.1.5 receptor-binding domain protects mice from the JN.1 variant.EBioMedicine. 2025 Jul;117:105794. doi: 10.1016/j.ebiom.2025.105794. Epub 2025 Jun 6. EBioMedicine. 2025. PMID: 40482468 Free PMC article.
-
One-Year Monitoring of the Evolution of SARS-CoV-2 Omicron Subvariants Through Wastewater Analysis (Central Italy, August 2023-July 2024).Life (Basel). 2025 May 24;15(6):850. doi: 10.3390/life15060850. Life (Basel). 2025. PMID: 40566504 Free PMC article.
-
Monitoring SARS-CoV-2 variants in wastewater during periods of low clinical case surveillance in Ethiopia.mSphere. 2025 Jul 29:e0022925. doi: 10.1128/msphere.00229-25. Online ahead of print. mSphere. 2025. PMID: 40728295
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications.Viruses. 2025 Jul 15;17(7):985. doi: 10.3390/v17070985. Viruses. 2025. PMID: 40733602 Free PMC article. Review.
References
-
- World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. Mar 11, 2020. [cited 30 December 2024]. Available from: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re... .
-
- World Health Organization. Tracking SARS-CoV-2 variants. [cited 26 February 2024]. Available from: https://www.who.int/activities/tracking-SARS-CoV-2-variants .
-
- Government of India. COVID-19 state wise data. [cited 28 February, 2024] Available from: https://www.mygov.in/covid-19-archive .
-
- Negi SS, Sharma K, Bhargava A, Singh P. A comprehensive profile of SARS-CoV-2 variants spreading during the COVID-19 pandemic: a genomic characterization study from Chhattisgarh State, India. Arch Microbiol. 2024;206:68. - PubMed
-
- Sharma K, Aggarwala P, Gandhi D, Mathias A, Singh P, Sharma S, Negi SS, Bhargava A, Das P, Gaikwad U, Wankhede A, Behra A, Nagarkar NM. Comparative analysis of various clinical specimens in detection of SARS-CoV-2 using rRT-PCR in new and follow up cases of COVID-19 infection: Quest for the best choice. PLoS One. 2021;16:e0249408. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

