Amelioration of Liver Fibrosis via In Situ Hepatic Stellate Cell Conversion Through Co-Inhibition of TGF-β and GSK-3 Signalling
- PMID: 40575954
- PMCID: PMC12203458
- DOI: 10.1111/liv.70187
Amelioration of Liver Fibrosis via In Situ Hepatic Stellate Cell Conversion Through Co-Inhibition of TGF-β and GSK-3 Signalling
Abstract
Background and aims: Liver fibrosis, a progressive condition driven by chronic liver injury and excessive scar tissue formation, can lead to cirrhosis, a life-threatening disease. Activation of hepatic stellate cells (HSCs) is central to fibrosis progression, yet current therapies fail to halt or reverse this process. This study evaluated a combination therapy targeting HSCs to ameliorate fibrosis and promote liver repair.
Methods: A small molecule cocktail, SBCH (SB431542, a TGF-β inhibitor, and CHIR99021, a GSK-3 inhibitor), was tested in three fibrosis models: CCl4-induced, bile duct ligation (BDL) and non-alcoholic steatohepatitis (NASH) with diethylnitrosamine (DEN). Therapeutic effects were assessed using phenotypic analyses, in vivo tracing and single-cell RNA sequencing to uncover mechanisms.
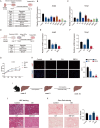
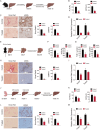
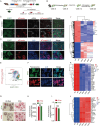
Results: SBCH significantly reduced fibrosis in all models by inhibiting HSC activation and fibrogenic activity. The suppression of PI3K/Akt pathway and EMT cascade contribute to the fibrosis-ameliorating effect of SBCH treatment. Furthermore, in vivo tracing and single-cell RNA sequencing revealed that SBCH induced the conversion of activated HSCs into hepatocyte-like cells (ciHeps), which integrated into liver tissue, repaired liver damage and restored liver integrity and function.
Conclusions: SBCH mitigates liver fibrosis through multifaceted mechanisms, including the inhibition of HSC activation, suppression of fibrogenic activity and regulation of key signalling pathways such as PI3K/Akt and EMT. In addition, SBCH induces the trans-differentiation of activated HSCs into hepatocyte-like cells (ciHeps), effectively reducing pathogenic HSCs while increasing functional ciHeps. This dual-target approach not only facilitates liver tissue repair but also restores liver function, offering a promising therapeutic strategy for liver fibrosis and cirrhosis, with potential applications in conditions arising from various aetiologies of liver injury.
Keywords: hepatic stellate cells; hepatocytes; lineage tracing; liver fibrosis and cirrhosis; trans‐differentiation.
© 2025 The Author(s). Liver International published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Xiao J., Wang F., Wong N.‐K., et al., “Global Liver Disease Burdens and Research Trends: Analysis From a Chinese Perspective,” Journal of Hepatology 71 (2019): 212–221. - PubMed
-
- Tsuchida T. and Friedman S. L., “Mechanisms of Hepatic Stellate Cell Activation,” Nature Reviews Gastroenterology & Hepatology 14 (2017): 397–411. - PubMed
-
- Younossi Z. M., Wong G., Anstee Q. M., and Henry L., “The Global Burden of Liver Disease,” Clinical Gastroenterology and Hepatology 21 (2023): 1978–1991. - PubMed
-
- Sung H., Ferlay J., Siegel R. L., et al., “Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries,” CA: A Cancer Journal for Clinicians 71 (2021): 209–249. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

