Single-Cell RNA Sequencing of Rabbit Sclera at Different Developmental Stages: Unveiling Scleral Cells Atlas and the Heterogeneity of Fibroblasts
- PMID: 40576432
- PMCID: PMC12212446
- DOI: 10.1167/iovs.66.6.83
Single-Cell RNA Sequencing of Rabbit Sclera at Different Developmental Stages: Unveiling Scleral Cells Atlas and the Heterogeneity of Fibroblasts
Abstract
Purpose: This study aims to construct a single-cell transcriptomic atlas of the developing rabbit sclera to elucidate fibroblast heterogeneity, differentiation trajectories, matrisome expression patterns, and intercellular communication, while revealing conserved molecular features of scleral cells through cross-species analysis.
Methods: Single-cell RNA sequencing (scRNA-seq) was performed on scleral tissues from New Zealand rabbits at embryonic day 25 (E25) and postnatal days 7 (P7), 21 (P21), and 180 (P180). Libraries were prepared using the DNBelab C Series Kit and sequenced on the BGISEQ-2000 platform. Sequencing reads were aligned to the OryCun2.0 genome using STAR, and unique molecular identifier (UMI) count matrices were generated with PISA. Data preprocessing was conducted using Seurat. Fibroblast lineage differentiation was analyzed via VIA, intercellular communication via CellChat, matrisome expression patterns via AUCell, and cross-species analyses via CACIMAR and hdWGCNA.
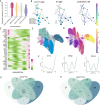
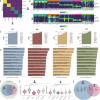
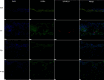
Results: We identified 7 major cell types and 15 subpopulations, with fibroblasts dominating the cellular landscape. Distinct fibroblast subtypes exhibited varied expression profiles and functions: KERAlow SPARCL1⁺ fibroblasts showed stem/progenitor-like features, while KERAhigh myocilin (MYOC)⁺ fibroblasts displayed senescence-associated phenotypes. Matrisome analysis revealed dynamic alterations in collagen and extracellular matrix (ECM)-related genes, and intercellular communication analysis highlighted complex signaling networks, particularly the MDK/PTN pathway. Cross-species comparisons demonstrated high conservation of fibroblasts between rabbit and human sclera, identifying four conserved co-expression modules.
Conclusions: This study presents the first single-cell atlas of rabbit scleral development, unveiling fibroblast heterogeneity, ECM remodeling mechanisms, and cross-species conserved features. These findings enhance our understanding of scleral biology and provide valuable insights for future research on ocular development and associated diseases, including myopia.
Conflict of interest statement
Disclosure:
Figures





References
-
- Gage PJ, Rhoades W, Prucka SK, Hjalt T.. Fate maps of neural crest and mesoderm in the mammalian eye. Invest Ophthalmol Vis Sci. 2005; 46(11): 4200–4208. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

