Genetically Engineered Yeast for Enhanced Biodegradation of Β-lactam Antibiotics
- PMID: 40576718
- PMCID: PMC12568803
- DOI: 10.1007/s12010-025-05291-4
Genetically Engineered Yeast for Enhanced Biodegradation of Β-lactam Antibiotics
Abstract
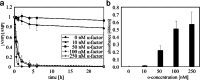
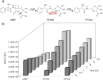
High environmental concentrations of pharmaceuticals, particularly antibiotics, have been observed worldwide, while antibiotic concentrations in the ng/L to µg/L range may adversely affect biota and contribute to the formation of antibiotic resistance. The underlying causes of high environmental concentrations include, in addition to the low rate of antibiotic' metabolization and the high rate of usage, especially of β-lactam antibiotics, the insufficient removal by conventional wastewater treatment methods. Consequently, alternative methods need to be developed to remove antibiotics from wastewater-one possibility is the use of enzymes. In this study, the enzyme β-lactamase was secreted by a genetically modified yeast (Saccharomyces cerevisiae) upon recognition of a pheromone (α-factor) as inducer to enable the degradation of ampicillin. This represents a crucial step on the road to a sensor-actuator system, allowing for the development of an intelligent removal system that can react to the presence of antibiotics. Ampicillin and its transformation products were studied by LC-MS/MS measurements using a carbamoyl functionalized column under hydrophilic interaction liquid chromatography (HILIC) conditions, which allowed detection of ampicillin at concentrations of 2.43 nM. The dependence of ampicillin degradation on α-factor concentration and the cultivation time of the yeast was demonstrated, resulting in higher degradation rate with higher α-factor concentrations and longer yeast cultivation times. Over 90% of 10 µM ampicillin was degraded within 0.5 h using 250 nM α-factor and a cultivation period of 24 h. Finally, the transferability to other β-lactam antibiotics was investigated, resulting in complete degradation of amoxicillin, penicillin G and piperacillin within 24 h.
Keywords: Antibiotics; Enzymatic degradation; LC–MS/MS; β-lactamase.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethic Approval: Not applicable. Consent to Participate: Not applicable. Consent for Publication: Not applicable. Conflict of interest: The authors declare no competing interests.
Figures




References
-
- Serwecińska, L. (2020). Antimicrobials and Antibiotic-Resistant Bacteria: A Risk to the Environment and to Public Health. Water,12(12), 3313.
-
- Klein, E. Y., Impalli, I., Poleon, S., Denoel, P., Cipriano, M., Van Boeckel, T. P., Pecetta, S., Bloom, D. E., & Nandi, A. (2024). Global trends in antibiotic consumption during 2016–2023 and future projections through 2030. Proceedings of the National Academy of Sciences,121(49), Article e2411919121. - PMC - PubMed
-
- Durand, G. A., Raoult, D., & Dubourg, G. (2019). Antibiotic discovery: History, methods and perspectives. International Journal of Antimicrobial Agents,53(4), 371–382. - PubMed
-
- Van Boeckel, T. P., Gandra, S., Ashok, A., Caudron, Q., Grenfell, B. T., Levin, S. A., & Laxminarayan, R. (2014). Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. The Lancet Infectious Diseases,14(8), 742–750. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

