Hepatitis B virus X protein (HBx)-mediated immune modulation and prognostic model development in hepatocellular carcinoma
- PMID: 40577282
- PMCID: PMC12204523
- DOI: 10.1371/journal.pone.0325363
Hepatitis B virus X protein (HBx)-mediated immune modulation and prognostic model development in hepatocellular carcinoma
Abstract
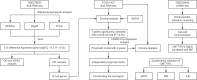
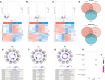
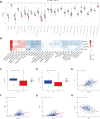
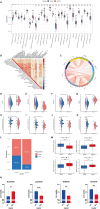
Hepatitis B virus (HBV) X protein (HBx) is critical in hepatocellular carcinoma (HCC) development, but its influence on tumor immunity and the tumor microenvironment (TME) remains unclear. This study aimed to construct a prognostic model based on HBx-related genes and explore their relationship with immune infiltration and immunotherapy response. Through transcriptome sequencing of our HBx-expressing HepG2 cells and analysis of HCC patient data from the cancer genome atlas (TCGA) and genotype-tissue expression (GTEx), we identified seven HBx-related genes, nuclear VCP-like (NVL), WD repeat domain 75 (WDR75), NOP58 nucleolar protein (NOP58), Brix domain-containing protein 1 (BRIX1), deoxynucleotidyltransferase terminal interacting protein 2 (DNTTIP2), MKI67 FHA domain interacting nucleolar phosphoprotein (NIFK), and ribosome production factor 2 (RPF2), associated with poor prognosis. LASSO Cox regression narrowed these to four key genes (BRIX1, RPF2, DNTTIP2, and WDR75), which were used to develop a prognostic riskscore signature. High-risk patients exhibited lower survival rates, decreased infiltration of anti-tumor immune cells, poorer responses to immunotherapy, and increased immune evasion. Among the four genes, DNTTIP2 showed higher expression in single-cell data, was linked to migration inhibitory factor (MIF) signaling, and may play a pivotal role in shaping an immunosuppressive TME. Elevated DNTTIP2 expression was confirmed in HBx-expressing HepG2 cells and HBV-infected HCC samples. This study highlights a novel HBx-related four-gene prognostic model that predicts clinical outcomes, immune infiltration, and immunotherapy response, offering insights into HCC progression and potential therapeutic targets.
Copyright: © 2025 Zhong et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

