Strain background interacts with chromosome 7 aneuploidy to determine commensal and virulence phenotypes in Candida albicans
- PMID: 40577316
- PMCID: PMC12204564
- DOI: 10.1371/journal.pgen.1011650
Strain background interacts with chromosome 7 aneuploidy to determine commensal and virulence phenotypes in Candida albicans
Abstract
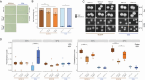
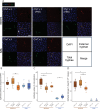
The human fungal pathobiont Candida albicans displays extensive genomic plasticity, including large-scale chromosomal changes such as aneuploidy. Chromosome trisomy appears frequently in natural and laboratory strains of C. albicans. Trisomy of specific chromosomes has been linked to large phenotypic effects, such as increased murine gut colonization by strains trisomic for chromosome 7 (Chr7). However, studies of whole-chromosome aneuploidy are generally limited to the SC5314 genome reference strain, making it unclear whether the imparted phenotypes are conserved across C. albicans genetic backgrounds. Here, we report the presence of a Chr7 trisomy in the "commensal-like" oral candidiasis strain, 529L, and dissect the contribution of Chr7 trisomy to colonization and virulence in 529L and SC5314. These experiments show that strain background and homolog identity (i.e., AAB vs ABB) interact with Chr7 trisomy to alter commensal and virulence phenotypes in multiple host niches. In vitro filamentation was consistently reduced by Chr7 trisomy in SC5314, but this result was not consistent for 529L. Oral colonization of mice was increased by the presence of a Chr7 trisomy in 529L but not SC5314; conversely, virulence during systemic infection was reduced by Chr7 trisomy in SC5314 but not 529L. Strikingly, the AAB Chr7 trisomy in the SC5314 background rendered this strain avirulent in murine systemic infection. Increased dosage of NRG1 failed to reproduce most of the Chr7 trisomy phenotypes. Our results demonstrate that aneuploidy interacts with background genetic variation to produce complex phenotypic patterns that deviate from our current understanding in the genome reference strain.
Copyright: © 2025 Mishra et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Update of
-
Strain background interacts with chromosome 7 aneuploidy to determine commensal and virulence phenotypes in Candida albicans.bioRxiv [Preprint]. 2025 Jan 24:2025.01.23.634449. doi: 10.1101/2025.01.23.634449. bioRxiv. 2025. Update in: PLoS Genet. 2025 Jun 27;21(6):e1011650. doi: 10.1371/journal.pgen.1011650. PMID: 39896449 Free PMC article. Updated. Preprint.
Similar articles
-
Strain background interacts with chromosome 7 aneuploidy to determine commensal and virulence phenotypes in Candida albicans.bioRxiv [Preprint]. 2025 Jan 24:2025.01.23.634449. doi: 10.1101/2025.01.23.634449. bioRxiv. 2025. Update in: PLoS Genet. 2025 Jun 27;21(6):e1011650. doi: 10.1371/journal.pgen.1011650. PMID: 39896449 Free PMC article. Updated. Preprint.
-
Commensal colonization of Candida albicans in the mouse gastrointestinal tract is mediated via expression of candidalysin and adhesins.Microbiol Spectr. 2025 Sep 2;13(9):e0056725. doi: 10.1128/spectrum.00567-25. Epub 2025 Jul 30. Microbiol Spectr. 2025. PMID: 40736264 Free PMC article.
-
Histone deacetylase Sir2 promotes the systemic Candida albicans infection by facilitating its immune escape via remodeling the cell wall and maintaining the metabolic activity.mBio. 2024 Jun 12;15(6):e0044524. doi: 10.1128/mbio.00445-24. Epub 2024 Apr 29. mBio. 2024. PMID: 38682948 Free PMC article.
-
Genomics-based non-invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women.Cochrane Database Syst Rev. 2017 Nov 10;11(11):CD011767. doi: 10.1002/14651858.CD011767.pub2. Cochrane Database Syst Rev. 2017. PMID: 29125628 Free PMC article.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
References
-
- Anderson FM, Visser ND, Amses KR, Hodgins-Davis A, Weber AM, Metzner KM, et al. Candida albicans selection for human commensalism results in substantial within-host diversity without decreasing fitness for invasive disease. PLoS Biol. 2023;21(5):e3001822. doi: 10.1371/journal.pbio.3001822 - DOI - PMC - PubMed

