Drosophila Trus, the orthologue of mammalian PDCD2L, is required for proper cell proliferation, larval developmental timing, and oogenesis
- PMID: 40577382
- PMCID: PMC12331172
- DOI: 10.1371/journal.pgen.1011469
Drosophila Trus, the orthologue of mammalian PDCD2L, is required for proper cell proliferation, larval developmental timing, and oogenesis
Abstract
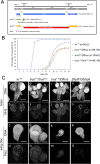
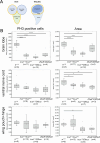
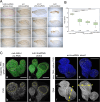
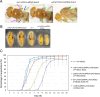
Toys are us (Trus) is the Drosophila melanogaster ortholog of mammalian Programmed Cell Death 2-Like (PDCD2L), a protein that has been implicated in ribosome biogenesis, cell cycle regulation, and oncogenesis. In this study, we examined the function of Trus during Drosophila development. CRISPR/Cas9 generated null mutations in trus lead to partial embryonic lethality, significant larval developmental delay, and complete pre-pupal lethality. In mutant larvae, we found decreased cell proliferation and growth defects in the brain and imaginal discs. Mapping relevant tissues for Trus function using trus RNAi and trus mutant rescue experiments revealed that imaginal disc defects are primarily responsible for the developmental delay, while the pre-pupal lethality is likely associated with faulty central nervous system (CNS) development. Examination of the molecular mechanism behind the developmental delay phenotype revealed that trus mutations induce the Xrp1-Dilp8 ribosomal stress-response in growth-impaired imaginal discs, and this signaling pathway attenuates production of the hormone ecdysone in the prothoracic gland. Additional Tap-tagging and mass spectrometry of components in Trus complexes isolated from Drosophila Kc cells identified Ribosomal protein subunit 2 (RpS2), which is coded by string of pearls (sop) in Drosophila, and Eukaryotic translation elongation factor 1 alpha 1 (eEF1α1) as interacting factors. We discuss the implication of these findings with respect to the similarity and differences in trus genetic null mutant phenotypes compared to the haplo-insufficiency phenotypes produced by heterozygosity for mutants in Minute genes and other genes involved in ribosome biogenesis.
Copyright: © 2025 Takada et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Update of
-
Drosophila Trus, the orthologue of mammalian PDCD2L, is required for proper cell proliferation, larval developmental timing, and oogenesis.bioRxiv [Preprint]. 2024 Oct 26:2024.10.24.620039. doi: 10.1101/2024.10.24.620039. bioRxiv. 2024. Update in: PLoS Genet. 2025 Jun 27;21(6):e1011469. doi: 10.1371/journal.pgen.1011469. PMID: 39484569 Free PMC article. Updated. Preprint.
References
-
- Bridges C, Morgan T. The third-chromosome group of mutant characters of Drosophila melanogaster. Carnegie Institute of Washington. 1923.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

