Viral hijacking of host DDX60 promotes Crimean-Congo haemorrhagic fever virus replication via G-quadruplex unwinding
- PMID: 40577441
- PMCID: PMC12221184
- DOI: 10.1371/journal.ppat.1013278
Viral hijacking of host DDX60 promotes Crimean-Congo haemorrhagic fever virus replication via G-quadruplex unwinding
Abstract
Crimean-Congo haemorrhagic fever virus (CCHFV) is the most prevalent tick-borne zoonotic bunyavirus, causing severe hemorrhagic fever and fatality in humans. Currently, the absence of approved vaccines or therapeutics for CCHFV infection necessitates the development of innovative therapeutic strategies. Here, we identify a guanine (G)-rich sequence located within the mRNA of the glycoprotein precursor in the medium (M) segment of the CCHFV genome, designated as M-PQS-1664(+). M-PQS-1664(+) can form stable G-quadruplex (G4) structure and functions as a negative regulatory element for viral replication. Host DDX60 is up-regulated in response to CCHFV infection, thereby it is hijacked to unwind M-PQS-1664(+) G4 for facilitating viral replication. The FDA-approved drug Cepharanthine (CEP), which competes with DDX60 to specifically stabilize M-PQS-1664(+) G4 without a global induction of host cellular G4s formation, exhibits remarkable antiviral activity in vitro and in vivo. More importantly, CEP possesses antiviral activity (50% inhibitory concentration ~ 0.2 μM) that having ~ 88 × the potency of ribavirin. Our findings underscore the CCHFV G4s as a promising target for drug development and highlight the significant potential of CEP in combating CCHFV.
Copyright: © 2025 Sui et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



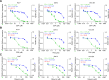

References
-
- Al-Abri SS, Abaidani IA, Fazlalipour M, Mostafavi E, Leblebicioglu H, Pshenichnaya N, et al. Current status of Crimean-Congo haemorrhagic fever in the World Health Organization Eastern Mediterranean Region: issues, challenges, and future directions. Int J Infect Dis. 2017;58:82–9. doi: 10.1016/j.ijid.2017.02.018 - DOI - PMC - PubMed
-
- Frank MG, Weaver G, Raabe V; State of the Clinical Science Working Group of the National Emerging Pathogens T, Education Center’s Special Pathogens Research N. Crimean-Congo Hemorrhagic Fever Virus for Clinicians-Epidemiology, Clinical Manifestations, and Prevention. Emerg Infect Dis. 2024;30:854–63. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

