Resolving the conformational ensemble of a membrane protein by integrating small-angle scattering with AlphaFold
- PMID: 40577488
- PMCID: PMC12251176
- DOI: 10.1371/journal.pcbi.1013187
Resolving the conformational ensemble of a membrane protein by integrating small-angle scattering with AlphaFold
Abstract
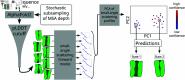
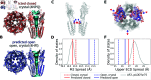
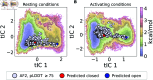
The function of a protein is enabled by its conformational landscape. For non-rigid proteins, a complete characterization of this landscape requires understanding the protein's structure in all functional states, the stability of these states under target conditions, and the transition pathways between them. Several strategies have recently been developed to drive the machine learning algorithm AlphaFold2 (AF) to sample multiple conformations, but it is more challenging to a priori predict what states are stabilized in particular conditions and how the transition occurs. Here, we combine AF sampling with small-angle scattering curves to obtain a weighted conformational ensemble of functional states under target environmental conditions. We apply this to the pentameric ion channel GLIC using small-angle neutron scattering (SANS) curves, and identify apparent closed and open states. By comparing experimental SANS data under resting and activating conditions, we can quantify the subpopulation of closed channels that open upon activation, matching both experiments and extensive simulation sampling using Markov state models. The predicted closed and open states closely resemble crystal structures determined under resting and activating conditions respectively, and project to predicted basins in free energy landscapes calculated from the Markov state models. Further, without using any structural information, the AF sampling also correctly captures intermediate conformations and projects onto the transition pathway resolved in the extensive sampling. This combination of machine learning algorithms and low-dimensional experimental data appears to provide an efficient way to predict not only stable conformations but also accurately sample the transition pathways several orders of magnitude faster than simulation-based sampling.
Copyright: © 2025 Lidbrink et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Werner C, Lindenblatt D, Viht K, Uri A, Niefind K. Discovery and exploration of protein kinase CK2 binding sites using CK2α′Cys336Ser as an exquisite crystallographic tool. Kinases Phosphatases. 2023;1(4):306–22. doi: 10.3390/kinasesphosphatases1040018 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

