FusionEncoder: identification of intrinsically disordered regions based on multi-feature fusion
- PMID: 40577786
- PMCID: PMC12231546
- DOI: 10.1093/bioinformatics/btaf362
FusionEncoder: identification of intrinsically disordered regions based on multi-feature fusion
Abstract
Motivation: Intrinsic disorder regions (IDRs) play a significant role in diverse biological processes and are widely distributed in proteins. Thus, accurately predicting these regions is essential for analyzing protein structure and function. Amino acid feature extraction servers as a foundational process in the development of computational predictive models. Existing methods typically rely on traditional biological features (e.g. PSSM) or use pre-trained protein language models (PPLMs) to capture sequence semantic information, often resorting to straightforward feature concatenation. However, these approaches fail to capture the multi-semantic interactions between traditional biological features and PPLMs-based features.
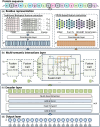
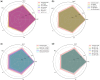
Results: In this study, we propose a method named FusionEncoder designed for the integration of traditional biological and PPLMs-based features of the protein. FusionEncoder is a fusion network built on a variant of long short-term memory (LSTM). We consider traditional biological features and PPLMs-based features to be two types of semantic inputs within a "multi-semantic" space. Traditional features are input into the cell state of the LSTM, while PPLMs-based features are fed into the input part. A fusion cell is then utilized to fuse these two types of features. This strategy leverages the capability of LSTM to encode long sequences, enhancing context-aware semantic learning of amino acid sequences. Finally, a transformer-based encoder layer is employed to predict the IDRs. Evaluation on four independent test datasets indicate that FusionEncoder obviously improves the accuracy of amino acid feature representation and achieves superior performance compared to the other existing methods.
Availability and implementation: To facilitate accessibility for experimental researchers, a user-friendly and publicly available webserver for the FusionEncoder predictor has been deployed at http://bliulab.net/FusionEncoder/. FusionEncoder is expected to serve as a valuable tool for the accurate identification of IDRs.
© The Author(s) 2025. Published by Oxford University Press.
Figures


References
-
- Bahdanau D, Cho KH, Bengio Y. Neural machine translation by jointly learning to align and translate. In: 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 2015.
-
- Chen J, Guo M, Li S et al. ProtDec-LTR2. 0: an improved method for protein remote homology detection by combining pseudo protein and supervised learning to rank. Bioinformatics 2017;33:3473–6. - PubMed
-
- Cheng H, Rao B, Liu L et al. PepFormer: end-to-end transformer-based siamese network to predict and enhance peptide detectability based on sequence only. Anal Chem 2021;93:6481–90. - PubMed
-
- Cheng J, Sweredoski MJ, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Min Knowl Discov 2005;11:213–22.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

