PHF6 and RUNX1 mutations cooperate to accelerate leukemogenesis
- PMID: 40577964
- PMCID: PMC12302341
- DOI: 10.1016/j.tranon.2025.102449
PHF6 and RUNX1 mutations cooperate to accelerate leukemogenesis
Abstract
Background: RUNX1 is a critical transcription factor in hematopoiesis and its mutations occur in various hematological diseases. PHF6 (plant homeodomain finger gene 6) is regarded as an epigenetic modifier, and its mutations are seen in myeloid and lymphoid leukemia. Previous studies have shown positive association of these two mutations. However, the joint pathological effects of these two genetic alterations remained unexplored.
Methods: We sought to investigate the pathological basis of the association between these two mutations. We first analyzed the clinical, genetic, and transcriptomic features of our cohort of patients with acute myeloid leuemia (AML) focusing on these two mutations. We transduced RUNX1 mutant into the genetically engineered Phf6 knockout (KO) mouse model to generate single- and double-mutated mice for in vivo experiments.
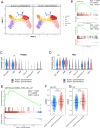
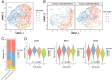
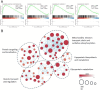
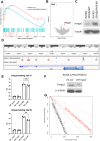
Results: In our 1188 adult AML patients, we observed frequent co-occurrence of PHF6 and RUNX1 mutations, and particularly worse clinical outcomes in these double-mutated patients. Double-mutated bone marrow (BM) cells displayed enriched leukemogenesis-related transcriptomic signatures and significantly higher engraftment capacity. The recipient mice transplanted with double-mutated BM cells developed AML with significantly shortened survival. Furthermore, we discovered that the multipotent progenitors (MPPs) were the main cell subpopulation responsible for double-mutated BM cell-induced leukemia. We noted significant up-regulation of high mobility group AT-hook 2 (Hmga2) in double-mutated MPPs and knock-down of Hmga2 abated the self-renewal capacity in vitro..
Conclusions: Our findings highlighted the synergistic leukemogenic potential of Phf6 and RUNX1 mutations in vivo, and provided insights into the molecular mechanisms accounting for this very high-risk disease entity.
Keywords: Acute myeloid leukemia; Hmga2; Leukemia stem cell; PHF6, RUNX1.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Miyagi S., Sroczynska P., Kato Y., Nakajima-Takagi Y., Oshima M., Rizq O., et al. The chromatin-binding protein Phf6 restricts the self-renewal of hematopoietic stem cells. Blood. 2019;133(23):2495–2506. Jun 6. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

