Adropin expression reflects circadian, lipoprotein, and mitochondrial processes in human tissues
- PMID: 40578684
- PMCID: PMC12274323
- DOI: 10.1016/j.molmet.2025.102196
Adropin expression reflects circadian, lipoprotein, and mitochondrial processes in human tissues
Abstract
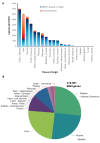
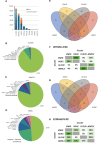
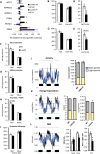
The clinical significance of interindividual variation in circulating adropin levels is unclear. To better understand adropin biology at the whole-body level, we surveyed transcriptional structures co-regulated with the Energy Homeostasis Associated (ENHO) gene encoding adropin across human tissues using Gene-Derived Correlations Across Tissues (GD-CAT). ENHO/adropin-related transcriptional structures with >1000 genes meeting the selection threshold (q < 0.001) occurred in 11/20 tissues. While most reflect local relationships, liver ENHO/adropin-related structures are dominated by transcripts expressed across metabolic tissues (skeletal muscle, adipose tissues, thyroid). Relationships between liver ENHO/adropin expression and skeletal muscle mitochondrial function were corroborated using liver-specific knockout mice. Within-liver ENHO/adropin transcriptional structures reflect lipoprotein metabolism (e.g., APOC1, p = 4.91 x 10-11; APOA1, p = 8.03 x 10-9), confirmed by correlations between plasma concentrations of adropin and indices of lipoprotein metabolism in MAPT samples. Moreover, statin treatment which increases hepatic cholesterol efflux, reduces plasma adropin levels. The ENHO gene contains retinoic acid receptor-related orphan receptor response elements (RORE), suggesting circadian control. Pan-organ transcriptional structures with liver ENHO/adropin or RORC overlap, reflecting the liver clock. Strong, local relationships between ENHO/adropin and circadian genes were also observed in most non-hepatic tissues. ENHO/adropin expression widely reflects activation of oxidative metabolic pathways and suppression of ribosomal functions and cell division. Finally, hippocampal ENHO/adropin expression correlates strongly with Alzheimer's disease risk genes identified by GWAS. In summary, activation of ENHO/adropin expression reflects cellular circadian and mitochondrial oxidative processes, but with inhibition of anabolic processes. Plasma adropin concentrations may thus reflect hepatic lipoprotein production and activation of metabolic stress responses across human tissues.
Copyright © 2025 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Andrew Butler reports financial support was provided by National Institutes of Health. Andrew Nguyen reports financial support was provided by National Institutes of Health. Marcus Seldin reports was provided by National Institutes of Health. Bruno Vellas reports was provided by French National Research Agency. Andrew Butler has patent #U.S. Patent no, 11/969,460, METHODS AND COMPOSITIONS FOR TREATING DECREASED COGNITIVE ABILITY issued to Saint Louis University. Nothing to declare If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Chen X., Chen S., Shen T., Yang W., Chen Q., Zhang P., et al. Adropin regulates hepatic glucose production via PP2A/AMPK pathway in insulin-resistant hepatocytes. FASEB J. 2020;34(8):10056–10072. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

