Distinct cytokine and cytokine receptor expression patterns characterize different forms of myositis
- PMID: 40579223
- PMCID: PMC12596072
- DOI: 10.1093/rheumatology/keaf346
Distinct cytokine and cytokine receptor expression patterns characterize different forms of myositis
Abstract
Objective: Myositis is a heterogeneous family of inflammatory myopathies. We sought to define the differential expression of cytokines, cytokine receptors and immune checkpoint genes in muscle biopsies from patients with different forms of myositis in order to characterize patterns of inflammation in each.
Methods: Bulk RNA-sequencing was performed on muscle biopsy samples from 669 patients, including 105 with DM, 80 with immune-mediated necrotizing myopathy (IMNM), 65 with anti-synthetase syndrome, 53 with IBM, 19 with anti-PM/Scl myositis, 310 with other inflammatory or genetic myopathies and 37 controls with normal tissue (NT). Myositis clinical groups and autoantibody subgroups were analysed separately. Expression data were analysed for 338 genes encoding cytokines, cytokine receptors and immune checkpoints. Myositis group-specific genes were identified from this list by finding genes that were specifically differentially expressed in one group compared with all samples and compared with NT (α < 0.001).
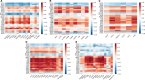
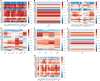
Results: IBM patients had the most differentially overexpressed genes (71) among all clinical groups, including 37 that were IBM-specific. Among the top genes were several involved in type 1 inflammation, including CCL5, CXCR3, CCR5, CXCL9 and IFNG. Anti-Jo1 and anti-PM/Scl patients exhibited differential overexpression of a similar set of genes, while DM patients exhibited differential overexpression of a different set of genes involved in type 1 inflammation. IMNM patients had the least number of differentially overexpressed genes with no predominant inflammatory pattern.
Conclusion: Each myositis clinical group and autoantibody subgroup had differentially overexpressed inflammatory mediators, including a strong type 1 inflammatory gene signature in IBM.
Keywords: DM; IBM; RNA-sequencing; cytokines; myositis.
Published by Oxford University Press on behalf of the British Society for Rheumatology 2025.
Figures

Update of
-
Distinct Cytokine and Cytokine Receptor Expression Patterns Characterize Different Forms of Myositis.medRxiv [Preprint]. 2025 Feb 21:2025.02.17.25321047. doi: 10.1101/2025.02.17.25321047. medRxiv. 2025. Update in: Rheumatology (Oxford). 2025 Nov 1;64(11):5751-5760. doi: 10.1093/rheumatology/keaf346. PMID: 40034760 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

