Circular RNA circIGF1R controls cardiac fibroblast proliferation through regulation of carbohydrate metabolism
- PMID: 40579400
- PMCID: PMC12205068
- DOI: 10.1038/s41598-025-07167-3
Circular RNA circIGF1R controls cardiac fibroblast proliferation through regulation of carbohydrate metabolism
Abstract
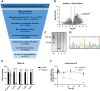
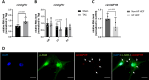
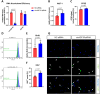
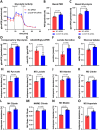
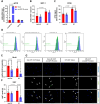
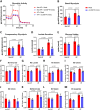
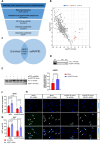
Excessive fibroblast proliferation and metabolic reprogramming are hallmarks of pathological cardiac remodeling, contributing significantly to impaired cardiac function. This study investigates the role of circular RNAs (circRNAs) in fibroblast metabolic reprogramming, an unexplored area with potential therapeutic implications. Through deep circRNA sequencing of cardiac tissue from heart failure (HF) patients and healthy individuals, we identified circIGF1R (hsa_circ_0005035), which exhibited dysregulation specifically in isolated cardiac fibroblasts derived from failing hearts. Silencing circIGF1R in patient-derived human cardiac fibroblasts (HCFs) led to accelerated proliferation, enhanced glycolytic activity, altered glucose trafficking, and increased glucose import. Conversely, administering recombinant circIGF1R inhibited the accelerated proliferation and enhanced glycolytic activity observed in HCFs from HF patients. Mechanistically, RNA pulldown assays and in silico analyses identified AZGP1 as a potential interaction partner facilitating the glycolysis-inhibitory and anti-proliferative functions of circIGF1R. Our findings identify circIGF1R as a pivotal regulator of fibroblast proliferation via metabolic reprogramming, particularly by glycolysis inhibition. Overexpression of circIGF1R demonstrated significant anti-fibrotic effects in cardiac fibroblasts derived from heart failure patients. These results underscore the therapeutic potential of circIGF1R in attenuating cardiac fibrosis by directly targeting fibroblast metabolism in the context of pathological cardiac remodeling.
Keywords: Cardiac fibroblast; Cardiac fibrosis; Cardiac metabolism; Circular RNA; Glycolysis; Heart failure; Proliferation.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval and consent to participate: The mice used in this study were purchased from Charles River. All animal experiments were approved by the authorities at Hannover Medical School and the Niedersächsische Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES). This study is performed in accordance with relevant guidelines and regulations. All methods are reported in accordance with ARRIVE guidelines. Ethical approval for the use of patient samples in this research was granted by the institutional ethics committee of the Hannover Medical School, Germany (number 9398_BO_K_2020). The study was conducted according to the guidelines from the declaration of Helsinki and its amendments or comparable ethical standards. Data collection of the patient samples including written informed consent was obtained.The ethics committee of the Hannover Medical School permitted and approved isolation of HCFs from patient’s biomaterial. Consent for publication: All authors have agreed to publish this manuscript.
Figures
References
-
- Nagaraju, C. K. et al. Myofibroblast phenotype and reversibility of fibrosis in patients with End-Stage heart failure. J. Am. Coll. Cardiol.73, 2267–2282 (2019). - PubMed
-
- Sun, Y. W. et al. Pirfenidone prevents radiation-induced intestinal fibrosis in rats by inhibiting fibroblast proliferation and differentiation and suppressing the TGF-beta1/Smad/CTGF signaling pathway. Eur. J. Pharmacol.822, 199–206 (2018). - PubMed
-
- Lewis, G. A. et al. Pirfenidone in heart failure with preserved ejection fraction: a randomized phase 2 trial. Nat. Med.27, 1477–1482 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

