A reference model of circulating hematopoietic stem cells across the lifespan with applications to diagnostics
- PMID: 40579545
- PMCID: PMC12283346
- DOI: 10.1038/s41591-025-03716-5
A reference model of circulating hematopoietic stem cells across the lifespan with applications to diagnostics
Abstract
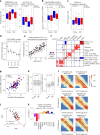
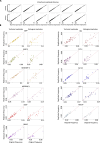
With aging, deviation of human blood counts from their normal range accompanies the transition from health to disease. Hematopoietic stem and progenitor cells (HSPCs) deliver life-long multi-lineage output, but their variation across healthy humans with aging, and their diagnostic utility, haven't been characterized in depth thus far. To address this, we introduced an HSPC reference model using single-cell RNA profiling of circulating CD34+ HSPCs from 148 healthy age- and sex-diverse individuals. We characterized physiological circulating HSPC composition, showed that age-related myeloid bias is predominant in older men and defined age-related transcriptional signatures in lymphoid progenitors. We further demonstrated the potential of this resource to facilitate the diagnosis of myelodysplastic syndrome (MDS) from peripheral blood without bone marrow sampling, defining classes of patients with MDS and abnormal lymphocyte, basophil or granulocyte progenitor frequencies. Our resource provides insights into HSPC reference ranges across the lifespan and has the potential to facilitate the clinical applications of single-cell genomics in hematology.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Z.S., D.L., E.M. and G.Y. are all employees and shareholders of Ultima Genomics. L.S. is a shareholder of Sequentify. L.S. and A.T. are shareholders of Cliseq. The other authors declare no competing interests.
Figures












References
-
- Osgood, E. E. Normal hematologic standards. Arch. Intern. Med.56, 849–863 (1935).
-
- Cohen, N. M. et al. Personalized lab test models to quantify disease potentials in healthy individuals. Nat. Med.10.1038/S41591-021-01468-6 (2021). - PubMed
-
- Mende, N. & Laurenti, E. Hematopoietic stem and progenitor cells outside the bone marrow: where, when, and why. Exp. Hematol.104, 9–16 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

