Ultra-low-input cell-free DNA sequencing for tumor detection and characterization in a real-world pediatric brain tumor cohort
- PMID: 40579709
- PMCID: PMC12205504
- DOI: 10.1186/s40478-025-02024-w
Ultra-low-input cell-free DNA sequencing for tumor detection and characterization in a real-world pediatric brain tumor cohort
Abstract
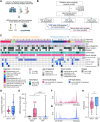
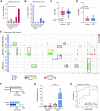
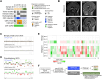
Molecular profiling of pediatric central nervous system (CNS) tumors has important clinical utility for guiding diagnostic and therapeutic strategies. Cell-free DNA (cfDNA) from liquid biopsies has been used for minimally invasive tumor profiling and longitudinal disease assessment in adult oncology and pediatric hematology. However, in pediatric neuro-oncology, low cfDNA yields pose a major barrier to translating these assays from bench to bedside. Here, we implemented a low-coverage whole genome sequencing (lcWGS) assay for picogram-level cfDNA inputs and applied it to liquid biopsies from a sizeable, population-based, cross-entity pediatric CNS tumor cohort (n = 56 patients). Applying this protocol, cfDNA whole genome profiles were successfully acquired from all liquid biopsy samples (n = 61/61 serum, n = 56/56 CSF, 100%). Based on copy number variations (CNVs), circulating-tumor DNA (ctDNA) was detected in 2/61 serum (3%) and in 25/56 CSF (45%) samples across various brain tumor entities. The integration of cfDNA results with clinical data demonstrated the utility of CSF lcWGS as a biomarker assay at diagnosis to distinguish cancerous from non-cancerous pineal region lesions (n = 6 patients). Additionally, serial CSF assessment in n = 9 patients (n = 29 CSF samples) enabled minimally invasive disease monitoring, with the added value of molecular profile availability in n = 4/6 (67%) patients at relapse. Proof-of-concept data show the feasibility of serial CSF lcWGS to reveal tumor evolution, tumor heterogeneity and potential therapeutic vulnerabilities in a case of medulloblastoma and germ cell tumor. Our study underscores the clinical utility of a robust lcWGS-based liquid biopsy assay optimized for low-input samples. We identify use-cases for implementing liquid biopsies in the clinical management of pediatric CNS tumor patients and provide a strong rationale for integration into future trials.
Keywords: Cell-free DNA; Cerebrospinal fluid liquid biopsies; Circulating-tumor DNA; Copy number variation profiling; Longitudinal molecular biomarkers; Low-coverage whole genome sequencing; Low-input samples; Minimally-invasive disease monitoring; Pediatric neuro-oncology; Tumor evolution.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Finnish Committee of Ethics of Tampere University Hospital (ethics approval ID: R13050). Written, informed consent was obtained from all patients and/or their legal representatives. Consent for publication: Written informed consent for the publication was obtained from all study participants, their parents, or legal guardians. Competing interests: S.M.P.: scientific advisory board at BioSkryb. M.S., S.M.P., and F.S.: co-founders and shareholders of Heidelberg Epignostix GmbH. MS has become full-time employee of Heidelberg Epignostix GmbH since July 2024.
Figures



References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72(1):7–33. 10.3322/caac.21708 - PubMed
-
- Ostrom QT, Price M, Ryan K, Edelson J, Neff C, Cioffi G et al (2022) CBTRUS statistical report: pediatric brain tumor foundation childhood and adolescent primary brain and other central nervous system tumors diagnosed in the united States in 2014–2018. Neuro Oncol 24(Suppl 3):iii1–iii38. 10.1093/neuonc/noac161 - PMC - PubMed
-
- Gröbner SN, Worst BC, Weischenfeldt J, Buchhalter I, Kleinheinz K, Rudneva VA et al (2018) The landscape of genomic alterations across childhood cancers. Nature 555(7696):321–327. 10.1038/nature25480 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

