N-Lactoyl-Phenylalanine modulates lipid metabolism in microglia/macrophage via the AMPK-PGC1α-PPARγ pathway to promote recovery in mice with spinal cord injury
- PMID: 40579710
- PMCID: PMC12205500
- DOI: 10.1186/s12974-025-03495-3
N-Lactoyl-Phenylalanine modulates lipid metabolism in microglia/macrophage via the AMPK-PGC1α-PPARγ pathway to promote recovery in mice with spinal cord injury
Abstract
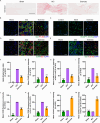
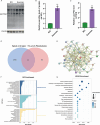
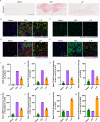
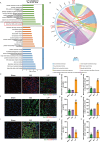
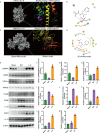
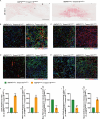
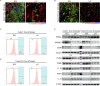
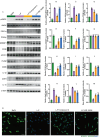
The accumulation of lipids in microglia/macrophage-induced inflammation exacerbation represents a pivotal factor contributing to secondary injury following spinal cord injury (SCI). N-Lactoyl-Phenylalanine (L-P), a metabolic byproduct of exercise, exhibits the capacity to regulate carbohydrate and lipid metabolism and may serve as a potential regulator of lipid metabolism in microglia/macrophage. This study investigates the role of L-P in modulating lipid homeostasis in microglia/macrophage and its therapeutic implications for SCI recovery. By establishing a mouse model of SCI, we confirmed that L-P administration markedly altered lipid metabolism in microglia/macrophage. This metabolic reprogramming was mediated through the activation of the AMPK-PGC1α-PPARγ signaling pathway, which plays a crucial role in regulating cellular energy metabolism and inflammatory responses. Our findings demonstrate that L-P treatment enhances the lipid metabolic capacity of microglia/macrophage, thereby attenuating neuroinflammation and promoting tissue repair after injury. Moreover, the polarization of microglia/macrophage shifts toward the anti-inflammatory M2 phenotype, providing substantial support for the regenerative process of the injured spinal cord. Functional analysis revealed that mice treated with L-P exhibited significantly improved motor function compared to the control group. Collectively, these results underscore the therapeutic potential of L-P in SCI and suggest its utility as a metabolic intervention strategy by modulating microglia/macrophage lipid metabolism to accelerate recovery.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics statement: The animal study was reviewed and approved by Animal Care and Use Committee of Wenzhou Medical College (Wydw2024–0539). Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Activation of the Nrf2 Signaling Pathway by Tetrahydroberberine Suppresses Ferroptosis and Enhances Functional Recovery Following Spinal Cord Injury.Mol Neurobiol. 2025 Jul;62(7):8439-8456. doi: 10.1007/s12035-025-04791-y. Epub 2025 Feb 26. Mol Neurobiol. 2025. PMID: 40011360
-
Interleukin-3 Modulates Macrophage Phagocytic Activity and Promotes Spinal Cord Injury Repair.CNS Neurosci Ther. 2024 Dec;30(12):e70181. doi: 10.1111/cns.70181. CNS Neurosci Ther. 2024. PMID: 39697159 Free PMC article.
-
[Effect of removing microglia from spinal cord on nerve repair after spinal cord injury in mice].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2025 Jun 15;39(6):754-761. doi: 10.7507/1002-1892.202503099. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2025. PMID: 40545466 Free PMC article. Chinese.
-
Phytochemicals regulate cancer metabolism through modulation of the AMPK/PGC-1α signaling pathway.BMC Cancer. 2024 Sep 2;24(1):1079. doi: 10.1186/s12885-024-12715-7. BMC Cancer. 2024. PMID: 39223494 Free PMC article.
-
Spatial and temporal activation of spinal glial cells: role of gliopathy in central neuropathic pain following spinal cord injury in rats.Exp Neurol. 2012 Apr;234(2):362-72. doi: 10.1016/j.expneurol.2011.10.010. Epub 2011 Oct 21. Exp Neurol. 2012. PMID: 22036747 Free PMC article.
References
-
- Troletti CD, Enzmann G, Chiurchiù V, Kamermans A, Tietz SM, Norris PC, Jahromi NH, Leuti A, van der Pol SMA, Schouten M, et al. Pro-resolving lipid mediator Lipoxin A attenuates neuro-inflammation by modulating T cell responses and modifies the spinal cord lipidome. Cell Rep. 2021;35:109201. - PMC - PubMed
-
- Li Y, Ritzel RM, Khan N, Cao TX, He JY, Lei ZF, Matyas JJ, Sabirzhanov B, Liu S, Li H, et al. Delayed microglial depletion after spinal cord injury reduces chronic inflammation and neurodegeneration in the brain and improves neurological recovery in male mice. Theranostics. 2020;10:11376–403. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

