DGHNN: a deep graph and hypergraph neural network for pan-cancer related gene prediction
- PMID: 40580449
- PMCID: PMC12254129
- DOI: 10.1093/bioinformatics/btaf379
DGHNN: a deep graph and hypergraph neural network for pan-cancer related gene prediction
Abstract
Motivation: Studies on pan-cancer related genes play important roles in cancer research and precision therapy. With the richness of research data and the development of neural networks, several successful methods that take advantage of multiomics data, protein interaction networks, and graph neural networks to predict cancer genes have emerged. However, these methods also have several problems, such as ignoring potentially useful biological data and providing limited representations of higher-order information.
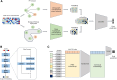
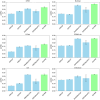
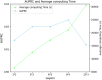
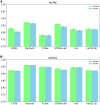
Results: In this work, we propose a pan-cancer related gene predictive model, the DGHNN, which takes biological pathways into consideration, applies a deep graph and hypergraph neural network to encode the higher-order information in the protein interaction network and biological pathway, introduces skip residual connections into the deep graph and hypergraph neural network to avoid problems with training the deep neural network, and finally uses a feature tokenizer and transformer for classification. The experimental results show that the DGHNN outperforms other methods and achieves state-of-the-art model performance for pan-cancer related gene prediction.
Availability and implementation: The DGHNN is available at https://github.com/skytea/DGHNN.
© The Author(s) 2025. Published by Oxford University Press.
Figures
References
-
- Bai S, Zhang F, Torr PHS. Hypergraph convolution and hypergraph attention. Pattern Recognit 2021;110:107637.
-
- Chen M, Wei Z, Huang Z et al. Simple and deep graph convolutional networks. In: Hal D III, Aarti S (eds.), Proceedings of the 37th International Conference on Machine Learning. Proceedings of Machine Learning Research: PMLR; 2020, 1725–35.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

