Development of the coronavirus reverse genetic system: Core technology for pathogenesis mechanisms research and vaccine/drug development
- PMID: 40580492
- PMCID: PMC12218529
- DOI: 10.1080/21505594.2025.2525930
Development of the coronavirus reverse genetic system: Core technology for pathogenesis mechanisms research and vaccine/drug development
Abstract
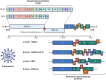
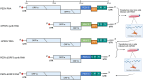
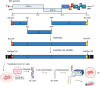
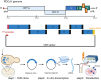
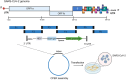
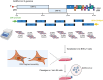
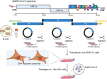
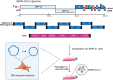
Coronaviruses (CoVs) are enveloped, single-stranded, positive-sense RNA viruses that cause respiratory, gastrointestinal, hepatic, and neurological diseases in humans and other animals. In recent years, frequent outbreaks of emerging and re-emerging CoVs have threatened animal and human health. However, an insufficient understanding of the mechanisms underlying CoV pathogenicity and cross-species transmission limits the development of drugs and vaccines against CoVs. Reverse genetic technology is a powerful tool for manipulating the genomes of CoVs and acquiring recombinant viruses, which allows researchers to better understand viral pathogenesis and develop genetically attenuated and marked vaccines and antiviral drugs. However, the large genomes of CoVs and the instability and toxicity of viral sequences in bacteria represent serious obstacles to the development of reverse genetic systems of CoVs. With the development of molecular biological methods, various new construction strategies have emerged. Accordingly, this review summarizes the construction strategies of CoV reverse genetics systems and their applications in studying pathogenesis, cross-species transmission, vaccine development, and drug screening, with the aim of providing an important reference for the prevention and control of CoVs.
Keywords: Coronavirus; antiviral drug-screening; coronavirus tropism; infectious clone; pathogenesis; reverse genetic system.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
A mutation in the coronavirus nsp13-helicase impairs enzymatic activity and confers partial remdesivir resistance.mBio. 2023 Aug 31;14(4):e0106023. doi: 10.1128/mbio.01060-23. Epub 2023 Jun 20. mBio. 2023. PMID: 37338298 Free PMC article.
-
Newly Emerged Antiviral Strategies for SARS-CoV-2: From Deciphering Viral Protein Structural Function to the Development of Vaccines, Antibodies, and Small Molecules.Int J Mol Sci. 2022 May 29;23(11):6083. doi: 10.3390/ijms23116083. Int J Mol Sci. 2022. PMID: 35682761 Free PMC article.
-
Rapid-response RNA-fluorescence in situ hybridization (FISH) assay platform for coronavirus antiviral high-throughput screening.SLAS Discov. 2024 Dec;29(8):100189. doi: 10.1016/j.slasd.2024.100189. Epub 2024 Nov 4. SLAS Discov. 2024. PMID: 39499968
-
Rapid reconstruction of infectious bronchitis virus expressing fluorescent protein from its nsp2 gene based on transformation-associated recombination platform.J Virol. 2025 Jul 22;99(7):e0053525. doi: 10.1128/jvi.00535-25. Epub 2025 Jun 5. J Virol. 2025. PMID: 40470960 Free PMC article.
-
COVID-19 Vaccines.2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. 2025 Jul 15. Drugs and Lactation Database (LactMed®) [Internet]. Bethesda (MD): National Institute of Child Health and Human Development; 2006–. PMID: 33355732 Free Books & Documents. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
