Comparative Analysis of Mammalian Adaptive Immune Loci Revealed Spectacular Divergence and Common Genetic Patterns
- PMID: 40580934
- PMCID: PMC12288031
- DOI: 10.1093/molbev/msaf152
Comparative Analysis of Mammalian Adaptive Immune Loci Revealed Spectacular Divergence and Common Genetic Patterns
Abstract
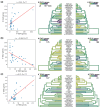
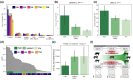
Adaptive immune responses are mediated by the production of adaptive immune receptors, antibodies, and T-cell receptors, which bind antigens, thus causing their neutralization. Unlike other proteins, adaptive immune receptors are not fully encoded in the germline genome and result from a complex of somatic processes collectively called V(D)J recombination affecting germline immunoglobulin (IG) and T-cell receptor (TR) loci consisting of template genes. While various existing studies report extreme diversity of antibodies and T-cell receptors, little is known about the diversity of germline IG and TR loci. To overcome this gap, the first comparative analysis of full-length sequences of IG/TR loci across 46 mammalian species from 13 taxonomic orders was performed. First, germline gene counts were shown to correlate in immunoglobulin heavy chain immunoglobulin heavy chain (IGH)/immunoglobulin lambda (IGL) loci and T-cell receptor alpha (TRA)/T-cell receptor beta (TRB) and anticorrelate in immunoglobulin kappa (IGK)/IGL, possibly indicating coevolution between corresponding chains. Second, structures of IG/TR loci were analyzed, and it was shown that IG/TR loci formed by long arrays of high multiplicity repeats are more common for species that have experienced population bottlenecks. Finally, haplotypes of IG/TR loci with little or no sequence similarity within a species were found, suggesting that they may have a limited potential for homologous recombination. These results demonstrate that IG/TR loci are rapidly evolving genomic regions whose structural variation is shaped by the population history of the species and open new perspectives for immunogenomics studies.
Keywords: T-cell receptor loci; adaptive immunity; genomics; immunogenomics; immunoglobulin loci; structural variations.
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: C.T.W. is a cofounder/CSO of Clareo Biosciences, Inc.
Figures



References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. The generation of antibody diversity. In Molecular biology of the cell. 4th ed. Garland Science; 2002. Chapter 24. https://www.ncbi.nlm.nih.gov/books/NBK21054/.
-
- Banach BB, Cerutti G, Fahad AS, Shen CH, De Souza MO, Katsamba PS, Tsybovsky Y, Wang P, Nair MS, Huang Y, et al. Paired heavy-and light-chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses. Cell Rep. 2021:37(1):109771. 10.1016/j.celrep.2021.109771. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

