Fitness Effect of the Isoniazid Resistance Mutation S315T of the Catalase-Peroxidase Enzyme KatG of Mycobacterium tuberculosis
- PMID: 40580943
- PMCID: PMC12242383
- DOI: 10.1093/gbe/evaf120
Fitness Effect of the Isoniazid Resistance Mutation S315T of the Catalase-Peroxidase Enzyme KatG of Mycobacterium tuberculosis
Abstract
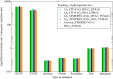
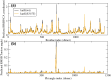
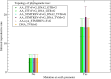
The mutation S315T of the catalase-peroxidase (CP) protein KatG of Mycobacterium tuberculosis is the most common mutation that confers resistance to the prodrug isoniazid. Here, we reconstruct its evolutionary history in 145 whole-genome sequences of M. tuberculosis from Russian hospitals, inferring 11 independent appearances of this mutation and 5 reversion events, with an estimated reversion rate 1,500 times higher than the rate of preserved nonsynonymous or intragenic mutations. This suggests that, contrary to the commonly held view, the mutation KatG(S315T) results in a fitness cost, possibly because of reduced tolerance to oxidative stress. Consistent with this interpretation, the mutant enzyme presents reduced CP activities. Applying the torsional network model (TNM), we found that the mutant protein shows more restricted thermal dynamics, although its functional site moves quite similarly to the wild type. Of the four internal clones where KatG(S315T) arose, two present high reproductive rates and secondary mutations at the 5'-UTR region of the gene encoding superoxide dismutase A (sodA), while the other two present significantly lower reproductive rates and lack mutations at genes related with tolerance to oxidative stress. Our results suggest that the resistance mutation KatG(S315T) incurs a fitness cost, which may be alleviated through compensatory mutations at the gene sodA or other genes that respond to oxidative stress, such as the previously known gene ahpC. This suggests that isoniazid treatment could be complemented with drugs that produce oxidative stress in order to hinder the propagation of resistant strains devoid of compensatory mutations.
Keywords: Mycobacterium tuberculosis; S315T; antimicrobial resistance; isoniazid; oxidative stress; regularized maximum likelihood and minimum evolution (REGMLAME).
© The Author(s) 2025. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest: None to be declared.
Figures




References
-
- Alfayate A, Rodriguez Caceres C, Gomes Dos Santos H, Bastolla U. Predicted dynamical couplings of protein residues characterize catalysis, transport and allostery. Bioinformatics. 2019:35:4971–4978. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

