The ability of SAMHD1-deficient monocytes to trigger the Type I IFN response depends on cGAS and mitochondrial DNA
- PMID: 40581121
- PMCID: PMC12312027
- DOI: 10.1016/j.jbc.2025.110430
The ability of SAMHD1-deficient monocytes to trigger the Type I IFN response depends on cGAS and mitochondrial DNA
Abstract
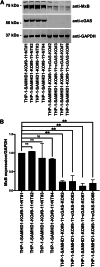
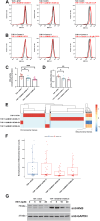
In humans, mutations in sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) lead to the development of a type I interferonopathy known as Aicardi-Goutières syndrome (AGS). AGS can present with a variety of severe phenotypes in patients, and a hallmark of this disease is chronic activation of type I interferon (IFN) signaling. However, the mechanism through which type I IFN signaling is activated in the absence of functional SAMHD1 is not known. Here, we investigated the molecular pathways that lead to type I IFN signaling activation in the absence of SAMHD1. Our investigations revealed that chronic activation of type I IFN signaling in SAMHD1-knockout (KO) monocytes is cyclic GMP-AMP synthase (cGAS)-dependent. Analysis of other nucleic acid sensors showed that type I IFN signaling in SAMHD1-KO cells is not dependent on melanoma differentiation-associated protein 5 (MDA5) or retinoic acid-inducible gene I (RIG-I). In agreement with our observation that type I IFN signaling is dependent on cGAS, two inhibitors of the cGAS-stimulator of IFN genes pathway, G140 and H151, effectively prevented type I IFN activation in SAMHD1-KO monocytes. We also found that type I IFN signaling in SAMHD1-KO monocytes is dependent on type I IFN receptor expression. Further exploration revealed mitochondrial malfunction in SAMHD1-KO monocytes that is likely to leak mitochondrial components into the cytoplasm. Overall, our work suggests that genetic knock out of SAMHD1 leads to mitochondrial disfunction, resulting in the presence of mitochondrial DNA in the cytoplasm, which triggers cGAS and the type I IFN response.
Keywords: Aicardi–Goutières syndrome; THP-1; cGAS; mitochondria; type I IFN response.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures









References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

