Fast and flexible minimizer digestion with digest
- PMID: 40581603
- PMCID: PMC12233085
- DOI: 10.1093/bioinformatics/btaf368
Fast and flexible minimizer digestion with digest
Abstract
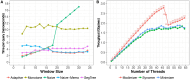
Summary: Minimizer digestion is an increasingly common component of bioinformatics tools, including tools for de Bruijn graph assembly and sequence classification. We describe a new open source tool and library to facilitate efficient digestion of genomic sequences. It can produce digests based on the related ideas of minimizers, modimizers or syncmers. Digest uses efficient data structures, scales well to many threads, and produces digests with expected spacings between digested elements.
Availability and implementation: Digest is implemented in C++17 with a Python API, and is available open-source at https://github.com/VeryAmazed/digest. The python library is available on Bioconda. Rust bindings are available as a public crate at https://crates.io/crates/digest-rs.
© The Author(s) 2025. Published by Oxford University Press.
Figures
Update of
-
Fast and flexible minimizer digestion with digest.bioRxiv [Preprint]. 2025 Jan 8:2025.01.02.631161. doi: 10.1101/2025.01.02.631161. bioRxiv. 2025. Update in: Bioinformatics. 2025 Jul 1;41(7):btaf368. doi: 10.1093/bioinformatics/btaf368. PMID: 39829865 Free PMC article. Updated. Preprint.
References
-
- Bentley JL. Solutions to Klee’s rectangle problems. Unpublished manuscript. 1977:282–300.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

