CXCR4 and CXCR6 dually limit T cell entry into the polyomavirus-infected brain
- PMID: 40581668
- PMCID: PMC12205503
- DOI: 10.1186/s12974-025-03496-2
CXCR4 and CXCR6 dually limit T cell entry into the polyomavirus-infected brain
Abstract
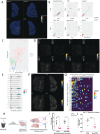
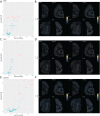
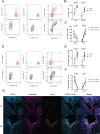
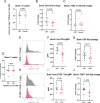
T cell responses are vital for controlling viral infection in the central nervous system (CNS), but must be tightly regulated to limit tissue-damaging inflammation. Using mouse polyomavirus (MuPyV) CNS infection, an in vivo model for JCPyV-induced Progressive Multifocal Leukoencephalopathy (PML), we investigated sites of early infection, immune responses, and recruitment of T cells to the brain. Multiplexed error-robust fluorescence in situ hybridization (MERFISH) single-cell spatial transcriptomics was applied to assess the regionality of virus infection and brain-resident cell and infiltrating leukocyte responses. MERFISH, immunofluorescence microscopy, quantitative PCR, and flow cytometry demonstrate that the ependyma is the predominant region of MuPyV CNS infection and localization of T cells, and implicated CXCR4 and CXCR6 in T cell migration to the ependyma and subventricular zone. Using CXCR6 knockout mice and a specific CXCR4 small molecule antagonist, we found that the combined impairment of CXCR6 and CXCR4 signaling resulted in elevated infiltration of T cells in the MuPyV-infected brain. This work demonstrates that CXCR4 and CXCR6 act in a nonredundant fashion to restrict T cell accumulation to the polyomavirus-infected ependyma, with important implications for ongoing efforts to use JCPyV-specific T cell adoptive immunotherapy for PML.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12974-025-03496-2.
Keywords: CXCR4; CXCR6; Ependyma; MuPyV; T cells.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
CD8 T Cells and STAT1 Signaling Are Essential Codeterminants in Protection from Polyomavirus Encephalopathy.J Virol. 2020 Mar 31;94(8):e02038-19. doi: 10.1128/JVI.02038-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996425 Free PMC article.
-
PD-1 Dynamically Regulates Inflammation and Development of Brain-Resident Memory CD8 T Cells During Persistent Viral Encephalitis.Front Immunol. 2019 Apr 17;10:783. doi: 10.3389/fimmu.2019.00783. eCollection 2019. Front Immunol. 2019. PMID: 31105690 Free PMC article.
-
CXCR6 promotes dermal CD8 + T cell survival and transition to long-term tissue residence.bioRxiv [Preprint]. 2025 May 30:2023.02.14.528487. doi: 10.1101/2023.02.14.528487. bioRxiv. 2025. PMID: 36824892 Free PMC article. Preprint.
-
Merkel cell carcinoma and Merkel cell polyomavirus: a systematic review and meta-analysis.Br J Dermatol. 2015 Jul;173(1):42-9. doi: 10.1111/bjd.13870. Epub 2015 Jun 12. Br J Dermatol. 2015. PMID: 25919492
-
Association between human polyomavirus infection and brain cancer: A systematic review and meta-analysis.Microb Pathog. 2022 Dec;173(Pt A):105797. doi: 10.1016/j.micpath.2022.105797. Epub 2022 Sep 29. Microb Pathog. 2022. PMID: 36183958
References
Grants and funding
LinkOut - more resources
Full Text Sources

