Calcitonin Gene-Related Peptide (CGRP)-Expressing Neurons in the External Lateral Parabrachial Area Regulate Pain-Induced Sleep Disturbances
- PMID: 40583282
- PMCID: PMC12463127
- DOI: 10.1002/advs.202500325
Calcitonin Gene-Related Peptide (CGRP)-Expressing Neurons in the External Lateral Parabrachial Area Regulate Pain-Induced Sleep Disturbances
Abstract
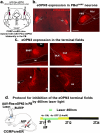
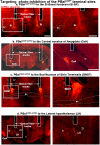
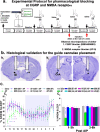
Given that sleep and pain are bidirectionally related, we investigated the neural circuits underlying pain-induced sleep disturbances using two acute pain models. Activation of nociceptors in acute inflammatory pain (AIP) significantly reduced sleep by 45-50% in the first 6 h, with reduced sleep spindle density for 1-3 h post-AIP. Additionally, an "optogenetic pain (Opto-Pain)" model is implemented to produce acute peripheral pain-induced awakenings that reduced sleep comparable to AIP. Both pain models are used to test the role of wake-promoting neurons in the parabrachial nucleus that express Calcitonin Gene-Related Peptide (PBelCGRP) in relaying nociceptive stimulus from the dorsal horn as part of the spine-ponto-amygdaloid tract. Blocking PBelCGRP neurons with genetic ablation or optogenetic inhibition attenuated sleep loss. To dissect the PBelCGRP pathways, the terminals are then optogenetically silenced post-AIP and found the reversal of sleep disturbances in the following descending order of effectiveness: substantia innominata of the basal forebrain (SI-BF) > central nucleus of the amygdala (CeA) > bed nucleus of the stria terminalis (BNST) > the lateral hypothalamus (LH). In SI-BF and CeA, a similar reversal of AIP-induced sleep loss occurred with pharmacological blocking of either CGRP or NMDA receptors. The results are relevant to emerging pain therapies aiming to attenuate sleep disturbances.
Keywords: acute pain; neural‐circuits; optogenetics; parabrachial nucleus; sleep‐loss.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Yong R. J., Mullins P. M., Bhattacharyya N., Pain 2022, 163, E328. - PubMed
-
- Sun Y., Laksono I., Selvanathan J., Saripella A., Nagappa M., Pham C., Englesakis M., Peng P., Morin C. M., Chung F., Sleep Med. Rev. 2021, 57, 101467. - PubMed
-
- Bonvanie I. J., Oldehinkel A. J., Rosmalen J. G. M., Janssens K. A. M., Pain 2016, 157, 957. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
