Fecal Microbial Community Profiling Allows Discrimination of Phenotype and Treatment Response in Pediatric Crohn's Disease and Ulcerative Colitis-An International Meta-Analysis
- PMID: 40583454
- PMCID: PMC12455605
- DOI: 10.1093/ibd/izaf135
Fecal Microbial Community Profiling Allows Discrimination of Phenotype and Treatment Response in Pediatric Crohn's Disease and Ulcerative Colitis-An International Meta-Analysis
Erratum in
-
Correction to: Fecal Microbial Community Profiling Allows Discrimination of Phenotype and Treatment Response in Pediatric Crohn's Disease and Ulcerative Colitis-An International Meta-Analysis.Inflamm Bowel Dis. 2025 Oct 1;31(10):2953. doi: 10.1093/ibd/izaf200. Inflamm Bowel Dis. 2025. PMID: 40907961 Free PMC article. No abstract available.
Abstract
Background and aims: The pathophysiology of pediatric inflammatory bowel disease (PIBD), encompassing Crohn's disease (CD) and ulcerative colitis (UC), is not entirely understood. Dysregulation of the intestinal microbiome is recognized as both a disease-driving and a potential therapeutic target. This study aimed to systematically analyze gut microbiome compositions and its applicability as a biomarker for disease progress and treatment response.
Methods: Bibliographic and nucleotide databases were searched. Raw 16S-rRNA sequencing reads were subjected to a uniform downstream dada2/phyloseq pipeline to extract taxonomy, community structure, and abundance information. Patient metadata were extracted from publications, and study authors were contacted for further details if required.
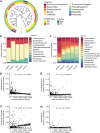
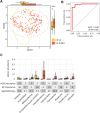
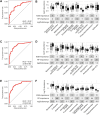
Results: Twenty-six studies comprising 3956 stool samples (CD 41%, UC 36%, 23% healthy) were included in the analyses. Median age of individuals was 12 (interquartile range 4). Sex distribution was comparable. Alpha diversity was reduced between the healthy and both UC and CD treatment-naïve groups (P < .001) and further reduced with increasing clinical disease activity. Beta diversity revealed altered community structure in treatment-naïve children with PIBD (P < .001). This alteration remained in patients in clinical remission (P < .001). Machine learning models discriminated between treatment-naïve patients with CD or UC with an area under the receiver operating characteristics curve (AUROC) of 98%. Microbial communities differed between patient responders versus nonresponders to treatment (P < .001). Further, microbial community profiling distinguished treatment response (eg, steroid, nutrition, or TNFα) with AUROCs of 82%-90%.
Conclusions: Gut microbial community structure is substantially altered in active and inactive PIBD and may be utilized as a biomarker for differentiating PIBD subtype and predicting treatment response.
Keywords: 16S metagenomics; Crohn’s disease; microbiome; treatment response; ulcerative colitis.
Plain language summary
We identified 26 studies on the gut microbiome in pediatric patients with IBD compiling a total of 3956 stool samples (CD 41%, UC 36%, 23% healthy) revealing microbial community structures unique to patients with CD and UC. These community patterns allow for the distinction of PIBD type and prediction of treatment response.
© 2025 Crohn’s & Colitis Foundation. Published by Oxford University Press on behalf of Crohn’s & Colitis Foundation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307-317. doi: https://doi.org/ 10.1038/nature10209 - DOI - PMC - PubMed
-
- Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: https://doi.org/ 10.1038/nmicrobiol.2017.4 - DOI - PMC - PubMed
-
- Dunne C. Adaptation of bacteria to the intestinal niche: probiotics and gut disorder. Inflamm Bowel Dis. 2001;7(2):136-145. doi: https://doi.org/ 10.1097/00054725-200105000-00010 - DOI - PubMed
-
- Radjabzadeh D, Boer CG, Beth SA, et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci Rep. 2020;10(1):1040. doi: https://doi.org/ 10.1038/s41598-020-57734-z - DOI - PMC - PubMed
-
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: https://doi.org/ 10.1371/journal.pbio.0050177 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

