The MpANT-Auxin Loop Modulates Marchantia polymorpha Development
- PMID: 40583831
- PMCID: PMC12207571
- DOI: 10.1111/ppl.70365
The MpANT-Auxin Loop Modulates Marchantia polymorpha Development
Abstract
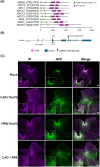
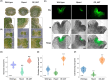
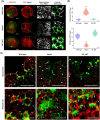
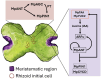
AINTEGUMENTA-LIKE/PLETHORA/BABYBOOM (APB) genes are considered part of the ancestral developmental toolkit in land plants. In Arabidopsis thaliana, these transcription factors are induced by auxin and are primarily expressed in tissues with actively dividing cells, where they play essential roles in organ development. Marchantia polymorpha, a liverwort that diverged from A. thaliana early in embryophyte evolution, possesses a single APB ortholog, MpAINTEGUMENTA (MpANT), encoded in its genome. In this study, we aimed to characterize the function of MpANT. Analysis of a transcriptional fusion line indicates that MpANT is predominantly expressed in the meristematic region. We report that the MpANT promoter region contains several cis-acting Auxin Responsive Elements (AREs) and demonstrate that its expression, which occurs predominantly in meristematic regions, is significantly altered by the addition of exogenous auxin and inhibition of auxin transport. These findings indicate that MpANT acts downstream of Auxin Response Factors (ARFs) and auxin signaling. Analyses of loss- and gain-of-function MpANT alleles highlight the importance of this transcription factor in meristem maintenance and cell proliferation. Additionally, we found that MpANT acts upstream of the auxin transporter MpPIN1 by influencing auxin distribution. Taken together, our findings reveal a feedforward regulatory loop involving auxin, MpANT, and MpPIN1 that is important for Marchantia development.
Keywords: Marchantia; ABP proteins; EvoDevo; PLETHORA; auxin.
© 2025 The Author(s). Physiologia Plantarum published by John Wiley & Sons Ltd on behalf of Scandinavian Plant Physiology Society.
Figures


References
-
- Aguilar‐Cruz, A. , Flores‐Sandoval E., Gutiérrez‐Ramos X., et al. 2023. “Control of Cell Fate Specification and Patterning by an Ancestral MicroRNA.” bioRxiv (Cold Spring Harbor Laboratory). 10.1101/2023.09.09.556951. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

