Decoding the Alphabet Soup: A Practical Guide to Genetic Testing in Hyperkinetic Movement Disorders
- PMID: 40584247
- PMCID: PMC12203903
- DOI: 10.5334/tohm.971
Decoding the Alphabet Soup: A Practical Guide to Genetic Testing in Hyperkinetic Movement Disorders
Abstract
Background: The diagnosis of genetic hyperkinetic movement disorders has become increasingly more complex as new genes are discovered and technologies offer new diagnostic possibilities. As a result, the choice of appropriate gene testing and the interpretation of the results can become difficult to navigate for movement disorder experts and clinicians. In parallel, research is becoming crucial to pair with clinical assessments in order to explore advanced sequencing technologies and allow new genes discovery.
Methods: Systematic review of genetic forms of hyperkinetic movement disorders and of the most relevant genetic terminology was performed.
Results: Comprehensive descriptions of genetic lexicon, testing selection, and complex genetic findings related to hyperkinetic movement disorders are reported.
Discussion: Here we discuss the terminology of genetic diagnosis that is now part of the clinical practice, the difficulties related to the interpretation of complex genetic results, and provide guidance and tips for gene testing selection in order not to miss important diagnosis of genetic hyperkinetic movement disorders.
Highlights: To review the most relevant lexicon related to genetic diagnosis, approach to gene testing, testing selection, and complex genetic findings in genetic hyperkinetic movement disorders.
Keywords: genetic tests; genotype-phenotype; hyperkinetic movement disorders; next generation sequencing; repeat expansion; variant of uncertain significance.
Copyright: © 2025 The Author(s).
Conflict of interest statement
The authors have no competing interests to declare.
Figures

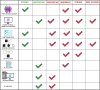
Similar articles
-
Scales for hyperkinetic disorders: A systematic review.J Neurol Sci. 2015 Nov 15;358(1-2):9-21. doi: 10.1016/j.jns.2015.08.1544. Epub 2015 Sep 2. J Neurol Sci. 2015. PMID: 26428309
-
Accidental Falls in Patients with Hyperkinetic Movement Disorders: A Systematic Review.Tremor Other Hyperkinet Mov (N Y). 2022 Oct 7;12:30. doi: 10.5334/tohm.709. eCollection 2022. Tremor Other Hyperkinet Mov (N Y). 2022. PMID: 36303814 Free PMC article.
-
Real-World Experiences with VMAT2 Inhibitors in Pediatric Hyperkinetic Movement Disorders.Tremor Other Hyperkinet Mov (N Y). 2025 Jun 16;15:26. doi: 10.5334/tohm.1023. eCollection 2025. Tremor Other Hyperkinet Mov (N Y). 2025. PMID: 40546692 Free PMC article.
-
Day 5 versus day 3 embryo biopsy for preimplantation genetic testing for monogenic/single gene defects.Cochrane Database Syst Rev. 2022 Nov 24;11(11):CD013233. doi: 10.1002/14651858.CD013233.pub2. Cochrane Database Syst Rev. 2022. PMID: 36423200 Free PMC article.
-
Idiopathic (Genetic) Generalized Epilepsy.2024 Feb 12. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Feb 12. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31536218 Free Books & Documents.
References
-
- Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med. 2020. Feb;22(2):245–57. DOI: 10.1038/s41436-019-0686-8 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

