Putative G‑Quadruplex Structures in Dysregulated Long Non-coding RNA of Ovarian Cancer and Their Binding Interactions with Human Serum Albumin
- PMID: 40584304
- PMCID: PMC12199190
- DOI: 10.1021/acsomega.5c00472
Putative G‑Quadruplex Structures in Dysregulated Long Non-coding RNA of Ovarian Cancer and Their Binding Interactions with Human Serum Albumin
Abstract
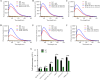
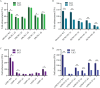
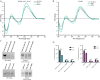
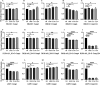
Long noncoding RNAs (lncRNAs) influence the progression, metastasis, and drug resistance of various cancers including ovarian cancer (OC). Putative G-quadruplex (G4)-forming sequences that are abundant in cancer-dysregulated lncRNAs have not been systematically pursued from a structure-function correlation perspective. In this work, we have used a combination of informatics, computational tools, spectroscopy, and molecular biology experiments to identify G4 formation by the OC-dysregulated lncRNAs ERLNC1, DLX6-AS1, LINC01127, FMNL1-DT, and LINP1. The in vitro ability of the lncRNAs to fold into G4 structures was accompanied by interesting profiles of individual G-tract contributions and response to monovalent cations, ligand TMPyP4, and G4-targeting antibody. Human serum albumin (HSA) was found to interact with these G4-forming lncRNAs, albeit with different affinities and structural implications for the G4 motifs. The G4-motif likely plays a crucial role in the binding interactions of select lncRNAs with HSA. This study provides the first systematic study of putative G4-forming sequences in OC-dysregulated lncRNAs and elucidates their interactions with HSA. The interaction of lncRNAs with HSA, possibly facilitated by G4 motifs, can be valuable for OC diagnosis and therapeutics.
© 2025 The Authors. Published by American Chemical Society.
Figures




Similar articles
-
Comparison of cellulose, modified cellulose and synthetic membranes in the haemodialysis of patients with end-stage renal disease.Cochrane Database Syst Rev. 2001;(3):CD003234. doi: 10.1002/14651858.CD003234. Cochrane Database Syst Rev. 2001. Update in: Cochrane Database Syst Rev. 2005 Jul 20;(3):CD003234. doi: 10.1002/14651858.CD003234.pub2. PMID: 11687058 Updated.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
LinkOut - more resources
Full Text Sources
