Simulations of Excitonic Couplings in Spinach Light-Harvesting Complex II Using a More Realistic Model in the Presence of Photosystem II Core
- PMID: 40584384
- PMCID: PMC12199089
- DOI: 10.1021/acsomega.4c11543
Simulations of Excitonic Couplings in Spinach Light-Harvesting Complex II Using a More Realistic Model in the Presence of Photosystem II Core
Abstract
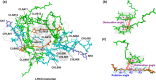
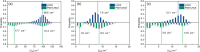
In this study, we employed a method for evaluating dynamic exciton couplings by using atomic transition density moments and molecular dynamics (MD) coordinates. We then applied this computational approach to investigate protein-regulated exciton couplings in spinach light-harvesting complex II (LHCII). Our findings, based on a more realistic all-atom computational model in the presence of the photosystem II (PSII) core and minor antennas, reveal distinct coupling strength patterns for the monomers in LHCII. The probability distributions of excitonic couplings in isolated LHCII always exhibit a single peak, whereas those in the LHCII-PSII complex display a bimodal nature. Such reductions in the excitonic coupling strengths of the main pigments, especially for some chlorophyll-lutein pairs, suggest that the peripheral proteins could modulate energy transfer processes in LHCII-PSII. A closer examination of the key structural parameters reveals that the angles between helices A and B and the distance between CLA612 and LUT620 increase in LHCII-PSII, which are closely associated with the reductions in excitonic couplings.
© 2025 The Authors. Published by American Chemical Society.
Figures









Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
Surgical interventions for treating extracapsular hip fractures in older adults: a network meta-analysis.Cochrane Database Syst Rev. 2022 Feb 10;2(2):CD013405. doi: 10.1002/14651858.CD013405.pub2. Cochrane Database Syst Rev. 2022. PMID: 35142366 Free PMC article.
-
Dynamic Coupling and Disorder in Aggregation of Light-Harvesting Complex II in Plant Thylakoid Membranes.J Phys Chem B. 2025 Sep 10. doi: 10.1021/acs.jpcb.5c05377. Online ahead of print. J Phys Chem B. 2025. PMID: 40928005
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Blankenship, R. E. Molecular mechanisms of photosynthesis, Blackwell Science, Oxford, 2002; pp 1–321.
-
- van Amerongen, H. ; van Grondelle, R. ; Valkunas, L. . Photosynthetic excitons; World Scientific, Singapore, 2000; pp 1–590.
LinkOut - more resources
Full Text Sources
