This is a preprint.
Multicenter Histology Image Integration and Multiscale Deep Learning for Machine Learning-Enabled Pediatric Sarcoma Classification
- PMID: 40585079
- PMCID: PMC12204438
- DOI: 10.1101/2025.06.10.25328700
Multicenter Histology Image Integration and Multiscale Deep Learning for Machine Learning-Enabled Pediatric Sarcoma Classification
Abstract
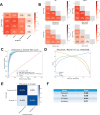
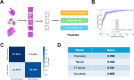
Pediatric sarcomas present diagnostic challenges due to their rarity and diverse subtypes, often requiring specialized pathology expertise and costly genetic tests. To overcome these barriers, we developed a computational pipeline leveraging deep learning methods to accurately classify pediatric sarcoma subtypes from digitized histology slides. To ensure classifier generalizability and minimize center-specific artifacts, we collected and harmonized a dataset comprising 867 whole slide images (WSIs) from three medical centers and the Children's Oncology Group (COG). Multiple convolutional neural network (CNN) and vision transformer (ViT) architectures were systematically evaluated as feature extractors for SAMPLER-based WSI representations, and input parameters such as tile size combinations and resolutions were tested and optimized. Our analysis showed that advanced ViT foundation models (UNI, CONCH) significantly outperformed earlier approaches, and incorporating multiscale features can enhance classification accuracy. Our optimized models achieved high performance, distinguishing rhabdomyosarcoma (RMS) from non-rhabdomyosarcoma (NRSTS) with an AUC of 0.969±0.026 and differentiating RMS subtypes (alveolar vs. embryonal) with an AUC of 0.961±0.021. Additionally, a two-stage pipeline effectively identified scarce Ewing sarcoma images from other NRSTS (AUC 0.929). Compared to conventional transformer encoder architectures used for WSI representations, our SAMPLER based classifiers were more lightweight (0.111 MB vs. 1.9 MB) and three orders of magnitude faster to train. This study highlights that digital histopathology paired with rigorous image harmonization provides a powerful solution for pediatric sarcoma classification. Our models reduce inter-observer variability, augment diagnostic precision, and have the potential to increase global accessibility to robust diagnostics, improving time to diagnosis and subsequent treatment planning.
Figures




References
-
- Children’s Oncology Group’s 2023. Blueprint for Research: Soft Tissue Sarcomas - PMC. https://pmc.ncbi.nlm.nih.gov/articles/PMC10519430/. - PMC - PubMed
-
- Rhabdomyosarcoma: Updates on classification and the necessity of molecular testing beyond immunohistochemistry - ClinicalKey. https://www.clinicalkey.com/#!/content/playContent/1-s2.0-S0046817723002.... - PubMed
-
- Williamson D. et al. Fusion Gene–Negative Alveolar Rhabdomyosarcoma Is Clinically and Molecularly Indistinguishable From Embryonal Rhabdomyosarcoma. J. Clin. Oncol. 28, 2151–2158 (2010). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
