MOLGENIS VIP: an end-to-end DNA variant interpretation pipeline for research and diagnostics configurable to support rapid implementation of new methods
- PMID: 40585303
- PMCID: PMC12205968
- DOI: 10.1093/nargab/lqaf087
MOLGENIS VIP: an end-to-end DNA variant interpretation pipeline for research and diagnostics configurable to support rapid implementation of new methods
Abstract
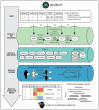
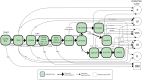
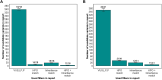
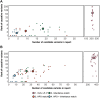
Achieving high yield in genetics research and genome diagnostics is a significant challenge because it requires a combination of multiple strategies and large-scale genomic analysis using the latest methods. Existing diagnostic software infrastructures are often unable to cope with high demands for versatility and scalability. We developed MOLGENIS VIP, a flexible, scalable, high-throughput, open-source, and "end-to-end" pipeline to process different types of sequencing data into portable, prioritized variant lists for immediate clinical interpretation in a wide variety of scenarios. VIP supports interpretation of short- and long-read sequencing data, using best-practice annotations and classification trees without complex IT infrastructures. VIP is developed within the long-living MOLGENIS open-source project to provide sustainability and has integrated feedback from a growing international community of users. VIP has undergone genome diagnostic laboratory testing and harnesses experiences from multiple Dutch, European, Canadian, and African diagnostic and infrastructural initiatives (VKGL, EU-Solve-RD, EJP-RD, CINECA, GA4GH). We provide a step-by-step protocol for installing and using VIP. We demonstrate VIP using 25 664 previously classified variants from the VKGL, and 18 and 41 diagnosed patients from a routine diagnostics and a Solve-RD research cohort, respectively. We believe that VIP accelerates causal variant detection and innovation in genome diagnostics and research.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Conflict of interest statement
None declared.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

