Differential cellular communication inference framework for large-scale single-cell RNA-sequencing data
- PMID: 40585304
- PMCID: PMC12204404
- DOI: 10.1093/nargab/lqaf084
Differential cellular communication inference framework for large-scale single-cell RNA-sequencing data
Abstract
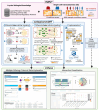
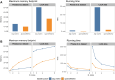
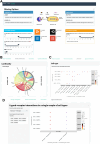
Single-cell transcriptomics data have been widely used to characterize biological systems, particularly in studying cell-cell communication, which plays a significant role in many biological processes. Despite the availability of various computational tools for inferring cellular communication, quantifying variations across different experimental conditions at both intercellular and intracellular levels remains challenging. Moreover, available methods are in general limited in terms of flexibility in analyzing different experimental designs and the ability to visualize results in an easily interpretable way. Here, we present a generalizable computational framework designed to infer and support differential cellular communication analysis across two experimental conditions from large-scale single-cell transcriptomics data. The scSeqCommDiff tool employs a statistical and network-based computational approach for characterizing altered cellular cross-talk in a fast and memory-efficient way. The framework is complemented with CClens, a user-friendly Shiny app to facilitate interactive analysis of inferred cell-cell communication. Validation through spatial transcriptomics data, comparison with other tools, and application to large-scale datasets (including a cell atlas) confirms the reliability, scalability, and efficiency of the framework. Moreover, the application to a single-nucleus transcriptomics dataset shows the validity and ability of the proposed workflow to support and unravel alterations in cell-cell interactions among patients with amyotrophic lateral sclerosis and healthy subjects.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Conflict of interest statement
None declared.
Figures



Similar articles
-
CellChat for systematic analysis of cell-cell communication from single-cell transcriptomics.Nat Protoc. 2025 Jan;20(1):180-219. doi: 10.1038/s41596-024-01045-4. Epub 2024 Sep 16. Nat Protoc. 2025. PMID: 39289562
-
SAKit: An all-in-one analysis pipeline for identifying novel proteins resulting from variant events at both large and small scales.J Bioinform Comput Biol. 2024 Oct;22(5):2450022. doi: 10.1142/S0219720024500227. Epub 2024 Oct 1. J Bioinform Comput Biol. 2024. PMID: 39573833
-
ScInfeR: an efficient method for annotating cell types and sub-types in single-cell RNA-seq, ATAC-seq, and spatial omics.Brief Bioinform. 2025 May 1;26(3):bbaf253. doi: 10.1093/bib/bbaf253. Brief Bioinform. 2025. PMID: 40471991 Free PMC article.
-
Unraveling cell-cell communication with NicheNet by inferring active ligands from transcriptomics data.Nat Protoc. 2025 Jun;20(6):1439-1467. doi: 10.1038/s41596-024-01121-9. Epub 2025 Mar 4. Nat Protoc. 2025. PMID: 40038548 Review.
-
Advances and challenges in cell-cell communication inference: a comprehensive review of tools, resources, and future directions.Brief Bioinform. 2025 May 1;26(3):bbaf280. doi: 10.1093/bib/bbaf280. Brief Bioinform. 2025. PMID: 40536815 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

