A Microfluidic Strategy to Capture Antigen-Specific High-Affinity B Cells
- PMID: 40585897
- PMCID: PMC12203763
- DOI: 10.1002/anbr.202300101
A Microfluidic Strategy to Capture Antigen-Specific High-Affinity B Cells
Abstract
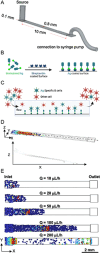
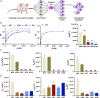
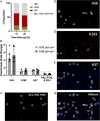
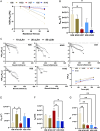
Assessing B cell affinity to pathogen-specific antigens prior to or following exposure could facilitate the assessment of immune status. Current standard tools to assess antigen-specific B cell responses focus on equilibrium binding of the secreted antibody in serum. These methods are costly, time-consuming, and assess antibody affinity under zero force. Recent findings indicate that force may influence BCR-antigen binding interactions and thus immune status. Herein, a simple laminar flow microfluidic chamber in which the antigen (hemagglutinin of influenza A) is bound to the chamber surface to assess antigen-specific BCR binding affinity of five hemagglutinin-specific hybridomas from 65 to 650 pN force range is designed. The results demonstrate that both increasing shear force and bound lifetime can be used to enrich antigen-specific high-affinity B cells. The affinity of the membrane-bound BCR in the flow chamber correlates well with the affinity of the matched antibodies measured in solution. These findings demonstrate that a microfluidic strategy can rapidly assess BCR-antigen-binding properties and identify antigen-specific high-affinity B cells. This strategy has the potential to both assess functional immune status from peripheral B cells and be a cost-effective way of identifying individual B cells as antibody sources for a range of clinical applications.
Keywords: affinities; avidities; cell separations; immunology.
© 2023 The Authors. Advanced NanoBiomed Research published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
A microfluidic strategy to capture antigen-specific high affinity B cells.bioRxiv [Preprint]. 2023 Jul 14:2023.07.12.548739. doi: 10.1101/2023.07.12.548739. bioRxiv. 2023. Update in: Adv Nanobiomed Res. 2024 Jun;4(6):2300101. doi: 10.1002/anbr.202300101. PMID: 37503139 Free PMC article. Updated. Preprint.
References
LinkOut - more resources
Full Text Sources
