OTUD1 inhibits osteoclast differentiation and osteoclastic bone loss through deubiquitinating and stabilizing PRDX1
- PMID: 40585986
- PMCID: PMC12203675
- DOI: 10.7150/thno.111360
OTUD1 inhibits osteoclast differentiation and osteoclastic bone loss through deubiquitinating and stabilizing PRDX1
Abstract
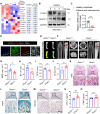
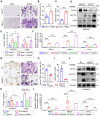
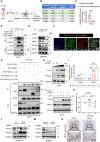
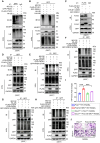
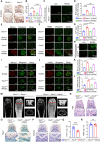
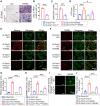
Rationale: Bone homeostasis relies on a delicate equilibrium between bone formation by osteoblasts and bone resorption by osteoclasts. Disruption of this balance leads to various disorders, most notably osteoporosis. Deubiquitinating enzymes (DUBs), which cleave ubiquitin moieties from substrate proteins, play critical regulatory roles in bone pathophysiology. In this study, we explored the function of a DUB, ovarian tumor deubiquitinase 1 (OTUD1), in bone remodeling. Methods: We examined the femur bone of Otud1+/+ and Otud1-/- male mice using micro-CT analyses and histomorphometry. The potential functions and mechanisms of OTUD1 were explored in bone marrow-derived macrophages, RAW264.7 cells, and bone marrow stromal cells using RT-qPCR, western blotting and immunofluorescence. Additionally, we employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) coupled with co-immunoprecipitation (Co-IP) to identify OTUD1-interacting proteins and substrates. Results: Our results demonstrated a significant downregulation of both the gene and protein level of OTUD1 during osteoclastogenesis. Furthermore, both whole-body knockout and myeloid-specific deficiency of OTUD1 resulted in reduced bone mass in male mice, driven by enhanced osteoclast differentiation. Mechanistically, OTUD1 maintained the stability of peroxiredoxin 1 (PRDX1) by reversing K48-linked ubiquitination, thereby mitigating mitochondrial dysfunction and suppressing osteoclast differentiation. Consistent with these results, mitochondria-targeted ubiquinone (MitoQ), a mitochondria-targeted antioxidant, effectively suppressed bone mass loss in OTUD1-deficient male mice. Conclusions: Our findings provided the first evidence that OTUD1 suppressed osteoclastogenesis by deubiquitinating PRDX1 and maintaining its stability, thereby offering a promising therapeutic approach for osteoclast-dependent bone diseases.
Keywords: Deubiquitination; Mitochondrial dysfunction; OTUD1; Osteoclast differentiation; PRDX1.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Veis DJ, O'Brien CA. Osteoclasts, master sculptors of bone. Annu Rev Pathol. 2023;18:257–81. - PubMed
-
- Liu J, Wu X, Lu J, Huang G, Dang L, Zhang H. et al. Exosomal transfer of osteoclast-derived miRNAs to chondrocytes contributes to osteoarthritis progression. Nat Aging. 2021;1:368–84. - PubMed
-
- Zheng J, Zhao J, Li C, Zhang F, Saiding Q, Zhang X. et al. Targeted protein fate modulating functional microunits promotes intervertebral fusion. Small Methods. 2024;8:e2301375. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

