Expanding automated multiconformer ligand modeling to macrocycles and fragments
- PMID: 40586518
- PMCID: PMC12208665
- DOI: 10.7554/eLife.103797
Expanding automated multiconformer ligand modeling to macrocycles and fragments
Abstract
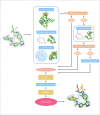
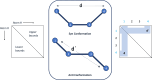
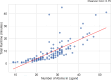
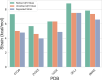
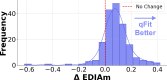
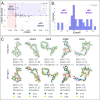
Small molecule ligands exhibit a diverse range of conformations in solution. Upon binding to a target protein, this conformational diversity is reduced. However, ligands can retain some degree of conformational flexibility even when bound to a receptor. In the Protein Data Bank, a small number of ligands have been modeled with distinct alternative conformations that are supported by macromolecular X-ray crystallography density maps. However, the vast majority of structural models are fit to a single-ligand conformation, potentially ignoring the underlying conformational heterogeneity present in the sample. We previously developed qFit-ligand to sample diverse ligand conformations and to select a parsimonious ensemble consistent with the density. While this approach indicated that many ligands populate alternative conformations, limitations in our sampling procedures often resulted in non-physical conformations and could not model complex ligands like macrocycles. Here, we introduce several improvements to qFit-ligand, including integrating RDKit for stochastic conformational sampling. This new sampling method greatly enriches low-energy conformations of small molecules and macrocycles. We further extended qFit-ligand to identify alternative conformations in PanDDA-modified density maps from high-throughput X-ray fragment screening experiments, as well as single-particle cryo-electron microscopy density maps. The new version of qFit-ligand improves fit to electron density and reduces torsional strain relative to deposited single-conformer models and our prior version of qFit-ligand. These advances enhance the analysis of residual conformational heterogeneity present in ligand-bound structures, which can provide important insights for the rational design of therapeutic agents.
Keywords: X-ray crystallography; compositional heterogeneity; conformational heterogeneity; cryo-EM; molecular biophysics; none; small molecules; structural biology.
© 2025, Flowers et al.
Conflict of interest statement
JF, NE, GC, PJ, TT, SW No competing interests declared, AR is a co-founder of TheRas, Elgia Therapeutics, and Tatara Therapeutics, and receives sponsored research support from Merck, Sharp and Dohme, Hv is an employee of Atomwise Inc, but the work in this publication does not overlap with his role there, JF is a consultant to, shareholder of, and receives sponsored research support from Relay Therapeutics and a consultant to and shareholder of Vilya Therapeutics
Figures










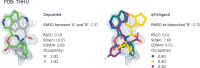

Update of
-
Expanding Automated Multiconformer Ligand Modeling to Macrocycles and Fragments.bioRxiv [Preprint]. 2025 Apr 30:2024.09.20.613996. doi: 10.1101/2024.09.20.613996. bioRxiv. 2025. Update in: Elife. 2025 Jun 30;14:RP103797. doi: 10.7554/eLife.103797. PMID: 39386683 Free PMC article. Updated. Preprint.
References
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica. Section D, Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
-
- Agrawal A, Verschueren R, Diamond S, Boyd S. A rewriting system for convex optimization problems. arXiv. 2017 doi: 10.48550/arXiv.1709.04494. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

