Minimization of proteome reallocation explains metabolic transition in hierarchical utilization of carbon sources
- PMID: 40586598
- PMCID: PMC12282068
- DOI: 10.1128/msystems.00690-25
Minimization of proteome reallocation explains metabolic transition in hierarchical utilization of carbon sources
Abstract
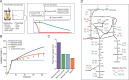
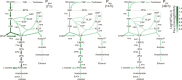
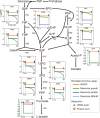
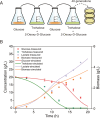
Cells choose between alternative pathways in metabolic networks under diverse environmental conditions, but the principles governing the choice are insufficiently understood, especially in response to dynamically changing conditions. Here, we observed that the lactic acid bacterium Bacillus coagulans displayed homolactic fermentation on glucose or trehalose as the sole carbon source but transitioned from homolactic to heterolactic fermentation during the hierarchical utilization of glucose and trehalose when growing on the mixture. We simulated the observation by dynamic minimization of reallocation of the proteome (dMORP) using an enzyme-constrained genome-scale metabolic model, which coincided with our omics data. Moreover, we evolved strains to co-utilize mixed carbon sources and repress the choice of heterolactic fermentation, and the dynamics after co-utilization of carbon sources were also captured by dMORP. Altogether, the findings suggest that upon environmental changes, bacteria tend to minimize proteome reallocation and accordingly adjust metabolism, and dMORP would be useful in simulating cellular dynamics.IMPORTANCERedundancy in metabolic networks empowers cells to choose between distinct metabolic strategies under changing environments. However, what drives the cellular choice remains poorly understood. We hypothesized that in response to rapid environmental changes, cells might minimize reallocation of the proteome and accordingly adjust metabolism. We found that this hypothesis could interpret a metabolic transition in the lactic acid bacterium Bacillus coagulans during the hierarchical utilization of glucose and trehalose, which was validated using systems biology approaches. Furthermore, we presented a framework with the objective function of minimizing proteome allocation, allowing for the simulation and understanding of cellular responses to dynamic perturbations.
Keywords: enzyme constraint; metabolic model; metabolic transition; mixed carbon sources.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Systems Biology Analysis of the Effect of pH on Lactate Fermentation in Bacillus coagulans Under Mixed Carbon Sources.Biotechnol Bioeng. 2025 Sep;122(9):2559-2573. doi: 10.1002/bit.70001. Epub 2025 Jun 12. Biotechnol Bioeng. 2025. PMID: 40503922
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
An integrated multiphase dynamic genome-scale model explains batch fermentations led by species of the Saccharomyces genus.mSystems. 2025 Feb 18;10(2):e0161524. doi: 10.1128/msystems.01615-24. Epub 2025 Jan 22. mSystems. 2025. PMID: 39840996 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Shen Y, Dinh HV, Cruz ER, Chen Z, Bartman CR, Xiao T, Call CM, Ryseck R-P, Pratas J, Weilandt D, Baron H, Subramanian A, Fatma Z, Wu Z-Y, Dwaraknath S, Hendry JI, Tran VG, Yang L, Yoshikuni Y, Zhao H, Maranas CD, Wühr M, Rabinowitz JD. 2024. Mitochondrial ATP generation is more proteome efficient than glycolysis. Nat Chem Biol 20:1123–1132. doi: 10.1038/s41589-024-01571-y - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
